Advancing microbiome science together: Welcome to the NMDC
We have built the National Microbiome Data Collaborative (NMDC) to advance how scientists create, use, and reuse data to redefine the way we understand and harness the power of microbes. The three core infrastructure elements of the NMDC framework are: (1) the Submission Portal to support collection of standardized study and biosample information; (2) NMDC EDGE, an intuitive user interface to access standardized bioinformatics workflows; and (3) the Data Portal, a resource for consistently processed and integrated multi-omics data enabling search, access, and download [2]. Our engagement strategy includes partnerships with complementary data resources like DOE’s Environmental Systems Science Data Infrastructure for a Virtual Ecosystem (ESS-DIVE) and DOE’s Systems Biology Knowledgebase (KBase); partnerships with DOE User Facilities, the Joint Genome Institute (JGI) and the Envionmental Molecular Sciences Laboratory (EMSL); coordinating with interagency programs outside of the DOE ecosystem such as NSF’s National Ecological Observatory Network (NEON); and development of our flagship engagement programs, the NMDC Ambassadors and Champions. This community-centric framework leverages unique capabilities, expertise, and resources available at the Department of Energy National Laboratories to create an enabling environment for findable, accessible, interoperable, and reusable (FAIR) multi-omics microbiome data.
About our Documentation
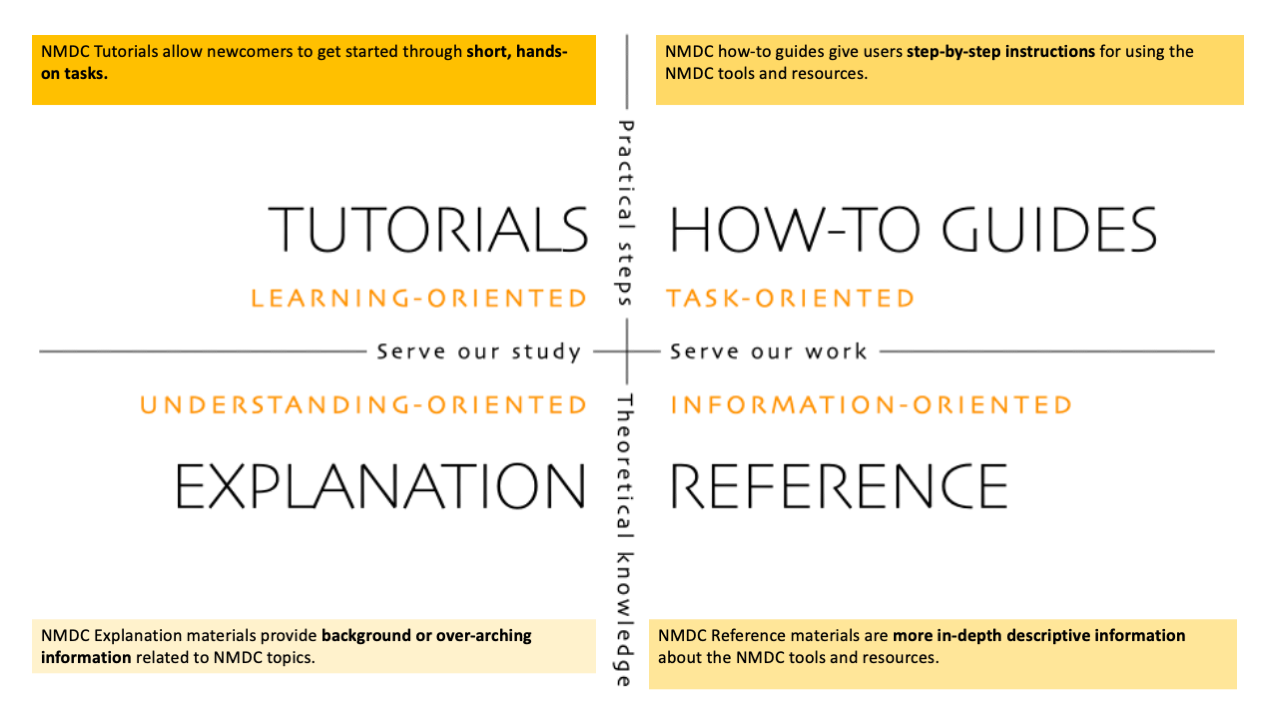
To support a systematic approach to our technical documentation, we have adopted the Diátaxis framework <https://diataxis.fr/> which identifies four distinct forms of documentation: tutorials, how-to guides, technical reference and explanation. We aim for our documentation to be accessible to a broad audience, so whether you are new to microbiome research or a leading scientist in the field, we welcome everyone to learn more from our:
Tutorials (Learning-oriented): get started through short, hands-on activities
How-To Guides (Task-oriented): step-by-step instructions for using NMDC’s products and resources
Explanation (Understanding-oriented): background information covering a wide range of topics
Reference (Information-oriented): in-depth learning materials
Let us know if you have suggestions to further support your research by reaching out to us!