NMDC Metadata Documentation
NMDC Schema
The purpose of the NMDC Schema is to define metadata for the National Microbiome Data Collaborative (NMDC). The NMDC is a multi-organizational effort to enable integrated microbiome data across diverse areas in medicine, agriculture, bioenergy, and the environment. This integrated platform facilitates comprehensive discovery of and access to multidisciplinary microbiome data in order to unlock new possibilities with microbiome data science.
The NMDC schema is used during the translation process to specify how metadata elements are related.
NMDC
metamodel version: 1.7.0
version: 2.1.0
Schema for National Microbiome Data Collaborative (NMDC).
This schema is organized into distinct modules:
a set of core types for representing data values
the mixs schema (auto-translated from mixs excel)
annotation schema
the NMDC schema itself
Classes
Activity - a provence-generating activity
WorkflowExecutionActivity - Represents an instance of an execution of a particular workflow
MetatranscriptomeActivity - A metatranscriptome activity that e.g. pools assembly and annotation activity.
Agent - a provence-generating agent
AttributeValue - The value for any value of a attribute for a sample. This object can hold both the un-normalized atomic value and the structured value
BooleanValue - A value that is a boolean
ControlledTermValue - A controlled term or class from an ontology
GeolocationValue - A normalized value for a location on the earth’s surface
ImageValue - An attribute value representing an image.
IntegerValue - A value that is an integer
PersonValue - An attribute value representing a person
QuantityValue - A simple quantity, e.g. 2cm
TextValue - A basic string value
TimestampValue - A value that is a timestamp. The range should be ISO-8601
UrlValue - A value that is a string that conforms to URL syntax
CreditAssociation - This class supports binding associated researchers to studies. There will be at least a slot for a CRediT Contributor Role (https://casrai.org/credit/) and for a person value Specifically see the associated researchers tab on the NMDC_SampleMetadata-V4_CommentsForUpdates at https://docs.google.com/spreadsheets/d/1INlBo5eoqn2efn4H2P2i8rwRBtnbDVTqXrochJEAPko/edit#gid=0
Database - An abstract holder for any set of metadata and data. It does not need to correspond to an actual managed databse top level holder class. When translated to JSON-Schema this is the ‘root’ object. It should contain pointers to other objects of interest
FunctionalAnnotation - An assignment of a function term (e.g. reaction or pathway) that is executed by a gene product, or which the gene product plays an active role in. Functional annotations can be assigned manually by curators, or automatically in workflows. In the context of NMDC, all function annotation is performed automatically, typically using HMM or Blast type methods
GenomeFeature - A feature localized to an interval along a genome
MetaboliteQuantification - This is used to link a metabolomics analysis workflow to a specific metabolite
NamedThing - a databased entity or concept/class
Biosample - A material sample. It may be environmental (encompassing many organisms) or isolate or tissue. An environmental sample containing genetic material from multiple individuals is commonly referred to as a biosample.
BiosampleProcessing - A process that takes one or more biosamples as inputs and generates one or as outputs. Examples of outputs include samples cultivated from another sample or data objects created by instruments runs.
OmicsProcessing - The methods and processes used to generate omics data from a biosample or organism.
DataObject - An object that primarily consists of symbols that represent information. Files, records, and omics data are examples of data objects.
GeneProduct - A molecule encoded by a gene that has an evolved function
Instrument - A material entity that is designed to perform a function in a scientific investigation, but is not a reagent[OBI].
-
ChemicalEntity - An atom or molecule that can be represented with a chemical formula. Include lipids, glycans, natural products, drugs. There may be different terms for distinct acid-base forms, protonation states
FunctionalAnnotationTerm - Abstract grouping class for any term/descriptor that can be applied to a functional unit of a genome (protein, ncRNA, complex).
OrthologyGroup - A set of genes or gene products in which all members are orthologous
Pathway - A pathway is a sequence of steps/reactions carried out by an organism or community of organisms
Reaction - An individual biochemical transformation carried out by a functional unit of an organism, in which a collection of substrates are transformed into a collection of products. Can also represent transporters
Person - represents a person, such as a researcher
Study - A study summarizes the overall goal of a research initiative and outlines the key objective of its underlying projects.
PeptideQuantification - This is used to link a metaproteomics analysis workflow to a specific peptide sequence and related information
ProteinQuantification - This is used to link a metaproteomics analysis workflow to a specific protein
ReactionParticipant - Instances of this link a reaction to a chemical entity participant
Mixins
Slots
INSDC identifiers - Any identifier covered by the International Nucleotide Sequence Database Collaboration
abstract - The abstract of manuscript/grant associated with the entity; i.e., a summary of the resource.
activity set - This property links a database object to the set of workflow activities.
add_date - The date on which the information was added to the database.
all proteins - the list of protein identifiers that are associated with the peptide sequence
protein quantification➞all proteins - the grouped list of protein identifiers associated with the peptide sequences that were grouped to a best protein
alternate emails - One or more other email addresses for an entity.
alternative descriptions - A list of alternative descriptions for the entity. The distinction between desciption and alternative descriptions is application-specific.
alternative identifiers - A list of alternative identifiers for the entity.
external database identifiers - Link to corresponding identifier in external database
GOLD sequencing project identifiers - identifiers for corresponding sequencing project in GOLD
-
GOLD analysis project identifiers - identifiers for corresponding analysis project in GOLD
-
GOLD sample identifiers - identifiers for corresponding sample in GOLD
INSDC biosample identifiers - identifiers for corresponding sample in INSDC
INSDC secondary sample identifiers - secondary identifiers for corresponding sample in INSDC
-
GOLD study identifiers - identifiers for corresponding project in GOLD
INSDC SRA ENA study identifiers - identifiers for corresponding project in INSDC SRA / ENA
INSDC bioproject identifiers - identifiers for corresponding project in INSDC Bioproject
MGnify project identifiers - identifiers for corresponding project in MGnify
alternative names - A list of alternative names used to refer to the entity. The distinction between name and alternative names is application-specific.
alternative titles - A list of alternative titles for the entity. The distinction between title and alternative titles is application-specific.
attribute - A attribute of a biosample. Examples: depth, habitat, material. For NMDC, attributes SHOULD be mapped to terms within a MIxS template
_16s_recover - Can a 16S gene be recovered from the submitted SAG or MAG?
_16s_recover_software - Tools used for 16S rRNA gene extraction
abs_air_humidity - Actual mass of water vapor - mh20 - present in the air water vapor mixture
adapters - Adapters provide priming sequences for both amplification and sequencing of the sample-library fragments. Both adapters should be reported; in uppercase letters
add_recov_method - Additional (i.e. Secondary, tertiary, etc.) recovery methods deployed for increase of hydrocarbon recovery from resource and start date for each one of them. If ‘other’ is specified, please propose entry in ‘additional info’ field
additional_info - Information that doesn’t fit anywhere else. Can also be used to propose new entries for fields with controlled vocabulary
address - The street name and building number where the sampling occurred.
adj_room - List of rooms (room number, room name) immediately adjacent to the sampling room
aero_struc - Aerospace structures typically consist of thin plates with stiffeners for the external surfaces, bulkheads and frames to support the shape and fasteners such as welds, rivets, screws and bolts to hold the components together
agrochem_addition - Addition of fertilizers, pesticides, etc. - amount and time of applications
air_temp - Temperature of the air at the time of sampling
air_temp_regm - Information about treatment involving an exposure to varying temperatures; should include the temperature, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include different temperature regimens
al_sat - Aluminum saturation (esp. For tropical soils)
al_sat_meth - Reference or method used in determining Al saturation
alkalinity - Alkalinity, the ability of a solution to neutralize acids to the equivalence point of carbonate or bicarbonate
alkalinity_method - Method used for alkalinity measurement
alkyl_diethers - Concentration of alkyl diethers
alt - Altitude is a term used to identify heights of objects such as airplanes, space shuttles, rockets, atmospheric balloons and heights of places such as atmospheric layers and clouds. It is used to measure the height of an object which is above the earthbs surface. In this context, the altitude measurement is the vertical distance between the earth’s surface above sea level and the sampled position in the air
aminopept_act - Measurement of aminopeptidase activity
ammonium - Concentration of ammonium in the sample
amniotic_fluid_color - Specification of the color of the amniotic fluid sample
amount_light - The unit of illuminance and luminous emittance, measuring luminous flux per unit area
ances_data - Information about either pedigree or other ancestral information description (e.g. parental variety in case of mutant or selection), e.g. A/3*B (meaning [(A x B) x B] x B)
annot - Tool used for annotation, or for cases where annotation was provided by a community jamboree or model organism database rather than by a specific submitter
annual_precpt - The average of all annual precipitation values known, or an estimated equivalent value derived by such methods as regional indexes or Isohyetal maps.
annual_temp - Mean annual temperature
antibiotic_regm - Information about treatment involving antibiotic administration; should include the name of antibiotic, amount administered, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple antibiotic regimens
api - API gravity is a measure of how heavy or light a petroleum liquid is compared to water (source: https://en.wikipedia.org/wiki/API_gravity) (e.g. 31.1B0 API)
arch_struc - An architectural structure is a human-made, free-standing, immobile outdoor construction
aromatics_pc - Saturate, Aromatic, Resin and AsphalteneB (SARA) is an analysis method that dividesB crude oilB components according to their polarizability and polarity. There are three main methods to obtain SARA results. The most popular one is known as the Iatroscan TLC-FID and is referred to as IP-143 (source: https://en.wikipedia.org/wiki/Saturate,_aromatic,_resin_and_asphaltene)
asphaltenes_pc - Saturate, Aromatic, Resin and AsphalteneB (SARA) is an analysis method that dividesB crude oilB components according to their polarizability and polarity. There are three main methods to obtain SARA results. The most popular one is known as the Iatroscan TLC-FID and is referred to as IP-143 (source: https://en.wikipedia.org/wiki/Saturate,_aromatic,_resin_and_asphaltene)
assembly_name - Name/version of the assembly provided by the submitter that is used in the genome browsers and in the community
assembly_qual - The assembly quality category is based on sets of criteria outlined for each assembly quality category. For MISAG/MIMAG; Finished: Single, validated, contiguous sequence per replicon without gaps or ambiguities with a consensus error rate equivalent to Q50 or better. High Quality Draft:Multiple fragments where gaps span repetitive regions. Presence of the 23S, 16S and 5S rRNA genes and at least 18 tRNAs. Medium Quality Draft:Many fragments with little to no review of assembly other than reporting of standard assembly statistics. Low Quality Draft:Many fragments with little to no review of assembly other than reporting of standard assembly statistics. Assembly statistics include, but are not limited to total assembly size, number of contigs, contig N50/L50, and maximum contig length. For MIUVIG; Finished: Single, validated, contiguous sequence per replicon without gaps or ambiguities, with extensive manual review and editing to annotate putative gene functions and transcriptional units. High-quality draft genome: One or multiple fragments, totaling 3 90% of the expected genome or replicon sequence or predicted complete. Genome fragment(s): One or multiple fragments, totalling < 90% of the expected genome or replicon sequence, or for which no genome size could be estimated
assembly_software - Tool(s) used for assembly, including version number and parameters
atmospheric_data - Measurement of atmospheric data; can include multiple data
avg_dew_point - The average of dew point measures taken at the beginning of every hour over a 24 hour period on the sampling day
avg_occup - Daily average occupancy of room
avg_temp - The average of temperatures taken at the beginning of every hour over a 24 hour period on the sampling day
bac_prod - Bacterial production in the water column measured by isotope uptake
bac_resp - Measurement of bacterial respiration in the water column
bacteria_carb_prod - Measurement of bacterial carbon production
barometric_press - Force per unit area exerted against a surface by the weight of air above that surface
basin - Name of the basin (e.g. Campos)
bathroom_count - The number of bathrooms in the building
bedroom_count - The number of bedrooms in the building
benzene - Concentration of benzene in the sample
bin_param - The parameters that have been applied during the extraction of genomes from metagenomic datasets
bin_software - Tool(s) used for the extraction of genomes from metagenomic datasets
biochem_oxygen_dem - Amount of dissolved oxygen needed by aerobic biological organisms in a body of water to break down organic material present in a given water sample at certain temperature over a specific time period
biocide - List of biocides (commercial name of product and supplier) and date of administration
biocide_admin_method - Method of biocide administration (dose, frequency, duration, time elapsed between last biociding and sampling) (e.g. 150 mg/l; weekly; 4 hr; 3 days)
biol_stat - The level of genome modification
biomass - Amount of biomass; should include the name for the part of biomass measured, e.g. Microbial, total. Can include multiple measurements
biotic_regm - Information about treatment(s) involving use of biotic factors, such as bacteria, viruses or fungi.
biotic_relationship - Description of relationship(s) between the subject organism and other organism(s) it is associated with. E.g., parasite on species X; mutualist with species Y. The target organism is the subject of the relationship, and the other organism(s) is the object
birth_control - Specification of birth control medication used
bishomohopanol - Concentration of bishomohopanol
blood_blood_disord - History of blood disorders; can include multiple disorders
bromide - Concentration of bromide
build_docs - The building design, construction and operation documents
build_occup_type - The primary function for which a building or discrete part of a building is intended to be used
building_setting - A location (geography) where a building is set
built_struc_age - The age of the built structure since construction
built_struc_set - The characterization of the location of the built structure as high or low human density
built_struc_type - A physical structure that is a body or assemblage of bodies in space to form a system capable of supporting loads
calcium - Concentration of calcium in the sample
carb_dioxide - Carbon dioxide (gas) amount or concentration at the time of sampling
carb_monoxide - Carbon monoxide (gas) amount or concentration at the time of sampling
carb_nitro_ratio - Ratio of amount or concentrations of carbon to nitrogen
ceil_area - The area of the ceiling space within the room
ceil_cond - The physical condition of the ceiling at the time of sampling; photos or video preferred; use drawings to indicate location of damaged areas
ceil_finish_mat - The type of material used to finish a ceiling
ceil_struc - The construction format of the ceiling
ceil_texture - The feel, appearance, or consistency of a ceiling surface
ceil_thermal_mass - The ability of the ceiling to provide inertia against temperature fluctuations. Generally this means concrete that is exposed. A metal deck that supports a concrete slab will act thermally as long as it is exposed to room air flow
ceil_type - The type of ceiling according to the ceiling’s appearance or construction
ceil_water_mold - Signs of the presence of mold or mildew on the ceiling
chem_administration - List of chemical compounds administered to the host or site where sampling occurred, and when (e.g. Antibiotics, n fertilizer, air filter); can include multiple compounds. For chemical entities of biological interest ontology (chebi) (v 163), http://purl.bioontology.org/ontology/chebi
chem_mutagen - Treatment involving use of mutagens; should include the name of mutagen, amount administered, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple mutagen regimens
chem_oxygen_dem - A measure of the capacity of water to consume oxygen during the decomposition of organic matter and the oxidation of inorganic chemicals such as ammonia and nitrite
chem_treatment - List of chemical compounds administered upstream the sampling location where sampling occurred (e.g. Glycols, H2S scavenger, corrosion and scale inhibitors, demulsifiers, and other production chemicals etc.). The commercial name of the product and name of the supplier should be provided. The date of administration should also be included
chem_treatment_method - Method of chemical administration(dose, frequency, duration, time elapsed between administration and sampling) (e.g. 50 mg/l; twice a week; 1 hr; 0 days)
chimera_check - A chimeric sequence, or chimera for short, is a sequence comprised of two or more phylogenetically distinct parent sequences. Chimeras are usually PCR artifacts thought to occur when a prematurely terminated amplicon reanneals to a foreign DNA strand and is copied to completion in the following PCR cycles. The point at which the chimeric sequence changes from one parent to the next is called the breakpoint or conversion point
chloride - Concentration of chloride in the sample
chlorophyll - Concentration of chlorophyll
climate_environment - Treatment involving an exposure to a particular climate; treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple climates
collection_date - The time of sampling, either as an instance (single point in time) or interval. In case no exact time is available, the date/time can be right truncated i.e. all of these are valid times: 2008-01-23T19:23:10+00:00; 2008-01-23T19:23:10; 2008-01-23; 2008-01; 2008; Except: 2008-01; 2008 all are ISO8601 compliant
compl_appr - The approach used to determine the completeness of a given SAG or MAG, which would typically make use of a set of conserved marker genes or a closely related reference genome. For UViG completeness, include reference genome or group used, and contig feature suggesting a complete genome
compl_score - Completeness score is typically based on either the fraction of markers found as compared to a database or the percent of a genome found as compared to a closely related reference genome. High Quality Draft: >90%, Medium Quality Draft: >50%, and Low Quality Draft: < 50% should have the indicated completeness scores
compl_software - Tools used for completion estimate, i.e. checkm, anvi’o, busco
conduc - Electrical conductivity of water
contam_score - The contamination score is based on the fraction of single-copy genes that are observed more than once in a query genome. The following scores are acceptable for; High Quality Draft: < 5%, Medium Quality Draft: < 10%, Low Quality Draft: < 10%. Contamination must be below 5% for a SAG or MAG to be deposited into any of the public databases
contam_screen_input - The type of sequence data used as input
contam_screen_param - Specific parameters used in the decontamination sofware, such as reference database, coverage, and kmers. Combinations of these parameters may also be used, i.e. kmer and coverage, or reference database and kmer
cool_syst_id - The cooling system identifier
crop_rotation - Whether or not crop is rotated, and if yes, rotation schedule
cult_root_med - Name or reference for the hydroponic or in vitro culture rooting medium; can be the name of a commonly used medium or reference to a specific medium, e.g. Murashige and Skoog medium. If the medium has not been formally published, use the rooting medium descriptors.
cur_land_use - Present state of sample site
cur_vegetation - Vegetation classification from one or more standard classification systems, or agricultural crop
cur_vegetation_meth - Reference or method used in vegetation classification
date_last_rain - The date of the last time it rained
decontam_software - Tool(s) used in contamination screening
density - Density of the sample, which is its mass per unit volume (aka volumetric mass density)
depos_env - Main depositional environment (https://en.wikipedia.org/wiki/Depositional_environment). If ‘other’ is specified, please propose entry in ‘additional info’ field
depth - Depth is defined as the vertical distance below local surface, e.g. For sediment or soil samples depth is measured from sediment or soil surface, respectively. Depth can be reported as an interval for subsurface samples
dermatology_disord - History of dermatology disorders; can include multiple disorders
detec_type - Type of UViG detection
dew_point - The temperature to which a given parcel of humid air must be cooled, at constant barometric pressure, for water vapor to condense into water.
diet_last_six_month - Specification of major diet changes in the last six months, if yes the change should be specified
diether_lipids - Concentration of diether lipids; can include multiple types of diether lipids
display order - When rendering information, this attribute to specify the order in which the information should be rendered.
diss_carb_dioxide - Concentration of dissolved carbon dioxide in the sample or liquid portion of the sample
diss_hydrogen - Concentration of dissolved hydrogen
diss_inorg_carb - Dissolved inorganic carbon concentration in the sample, typically measured after filtering the sample using a 0.45 micrometer filter
diss_inorg_nitro - Concentration of dissolved inorganic nitrogen
diss_inorg_phosp - Concentration of dissolved inorganic phosphorus in the sample
diss_iron - Concentration of dissolved iron in the sample
diss_org_carb - Concentration of dissolved organic carbon in the sample, liquid portion of the sample, or aqueous phase of the fluid
diss_org_nitro - Dissolved organic nitrogen concentration measured as; total dissolved nitrogen - NH4 - NO3 - NO2
diss_oxygen - Concentration of dissolved oxygen
diss_oxygen_fluid - Concentration of dissolved oxygen in the oil field produced fluids as it contributes to oxgen-corrosion and microbial activity (e.g. Mic).
-
study➞doi - The dataset citation for this study
dominant_hand - Dominant hand of the subject
door_comp_type - The composite type of the door
door_cond - The phsical condition of the door
door_direct - The direction the door opens
door_loc - The relative location of the door in the room
door_mat - The material the door is composed of
door_move - The type of movement of the door
door_size - The size of the door
door_type - The type of door material
door_type_metal - The type of metal door
door_type_wood - The type of wood door
door_water_mold - Signs of the presence of mold or mildew on a door
douche - Date of most recent douche
down_par - Visible waveband radiance and irradiance measurements in the water column
drainage_class - Drainage classification from a standard system such as the USDA system
drawings - The buildings architectural drawings; if design is chosen, indicate phase-conceptual, schematic, design development, and construction documents
drug_usage - Any drug used by subject and the frequency of usage; can include multiple drugs used
efficiency_percent - Percentage of volatile solids removed from the anaerobic digestor
elev - Elevation of the sampling site is its height above a fixed reference point, most commonly the mean sea level. Elevation is mainly used when referring to points on the earth’s surface, while altitude is used for points above the surface, such as an aircraft in flight or a spacecraft in orbit
elevator - The number of elevators within the built structure
emulsions - Amount or concentration of substances such as paints, adhesives, mayonnaise, hair colorants, emulsified oils, etc.; can include multiple emulsion types
encoded_traits - Should include key traits like antibiotic resistance or xenobiotic degradation phenotypes for plasmids, converting genes for phage
env_broad_scale - In this field, report which major environmental system your sample or specimen came from. The systems identified should have a coarse spatial grain, to provide the general environmental context of where the sampling was done (e.g. were you in the desert or a rainforest?). We recommend using subclasses of ENVOUs biome class: http://purl.obolibrary.org/obo/ENVO_00000428. Format (one term): termLabel [termID], Format (multiple terms): termLabel [termID]|termLabel [termID]|termLabel [termID]. Example: Annotating a water sample from the photic zone in middle of the Atlantic Ocean, consider: oceanic epipelagic zone biome [ENVO:01000033]. Example: Annotating a sample from the Amazon rainforest consider: tropical moist broadleaf forest biome [ENVO:01000228]. If needed, request new terms on the ENVO tracker, identified here: http://www.obofoundry.org/ontology/envo.html
env_local_scale - In this field, report the entity or entities which are in your sample or specimenUs local vicinity and which you believe have significant causal influences on your sample or specimen. Please use terms that are present in ENVO and which are of smaller spatial grain than your entry for env_broad_scale. Format (one term): termLabel [termID]; Format (multiple terms): termLabel [termID]|termLabel [termID]|termLabel [termID]. Example: Annotating a pooled sample taken from various vegetation layers in a forest consider: canopy [ENVO:00000047]|herb and fern layer [ENVO:01000337]|litter layer [ENVO:01000338]|understory [01000335]|shrub layer [ENVO:01000336]. If needed, request new terms on the ENVO tracker, identified here: http://www.obofoundry.org/ontology/envo.html
env_medium - In this field, report which environmental material or materials (pipe separated) immediately surrounded your sample or specimen prior to sampling, using one or more subclasses of ENVOUs environmental material class: http://purl.obolibrary.org/obo/ENVO_00010483. Format (one term): termLabel [termID]; Format (multiple terms): termLabel [termID]|termLabel [termID]|termLabel [termID]. Example: Annotating a fish swimming in the upper 100 m of the Atlantic Ocean, consider: ocean water [ENVO:00002151]. Example: Annotating a duck on a pond consider: pond water [ENVO:00002228]|air ENVO_00002005. If needed, request new terms on the ENVO tracker, identified here: http://www.obofoundry.org/ontology/envo.html
env_package - MIxS extension for reporting of measurements and observations obtained from one or more of the environments where the sample was obtained. All environmental packages listed here are further defined in separate subtables. By giving the name of the environmental package, a selection of fields can be made from the subtables and can be reported
escalator - The number of escalators within the built structure
estimated_size - The estimated size of the genome prior to sequencing. Of particular importance in the sequencing of (eukaryotic) genome which could remain in draft form for a long or unspecified period.
ethylbenzene - Concentration of ethylbenzene in the sample
execution resource - Example: NERSC-Cori
exp_duct - The amount of exposed ductwork in the room
exp_pipe - The number of exposed pipes in the room
experimental_factor - Experimental factors are essentially the variable aspects of an experiment design which can be used to describe an experiment, or set of experiments, in an increasingly detailed manner. This field accepts ontology terms from Experimental Factor Ontology (EFO) and/or Ontology for Biomedical Investigations (OBI). For a browser of EFO (v 2.95) terms, please see http://purl.bioontology.org/ontology/EFO; for a browser of OBI (v 2018-02-12) terms please see http://purl.bioontology.org/ontology/OBI
ext_door - The number of exterior doors in the built structure
ext_wall_orient - The orientation of the exterior wall
ext_window_orient - The compass direction the exterior window of the room is facing
extrachrom_elements - Do plasmids exist of significant phenotypic consequence (e.g. ones that determine virulence or antibiotic resistance). Megaplasmids? Other plasmids (borrelia has 15+ plasmids)
extreme_event - Unusual physical events that may have affected microbial populations
extreme_salinity - Measured salinity
fao_class - Soil classification from the FAO World Reference Database for Soil Resources. The list can be found at http://www.fao.org/nr/land/sols/soil/wrb-soil-maps/reference-groups
feat_pred - Method used to predict UViGs features such as ORFs, integration site, etc.
fertilizer_regm - Information about treatment involving the use of fertilizers; should include the name of fertilizer, amount administered, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple fertilizer regimens
field - Name of the hydrocarbon field (e.g. Albacora)
file size bytes - Size of the file in bytes
filter_type - A device which removes solid particulates or airborne molecular contaminants
fire - Historical and/or physical evidence of fire
fireplace_type - A firebox with chimney
flooding - Historical and/or physical evidence of flooding
floor_age - The time period since installment of the carpet or flooring
floor_area - The area of the floor space within the room
floor_cond - The physical condition of the floor at the time of sampling; photos or video preferred; use drawings to indicate location of damaged areas
floor_count - The number of floors in the building, including basements and mechanical penthouse
floor_finish_mat - The floor covering type; the finished surface that is walked on
floor_struc - Refers to the structural elements and subfloor upon which the finish flooring is installed
floor_thermal_mass - The ability of the floor to provide inertia against temperature fluctuations
floor_water_mold - Signs of the presence of mold or mildew in a room
fluor - Raw or converted fluorescence of water
foetal_health_stat - Specification of foetal health status, should also include abortion
freq_clean - The number of times the building is cleaned per week
freq_cook - The number of times a meal is cooked per week
fungicide_regm - Information about treatment involving use of fungicides; should include the name of fungicide, amount administered, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple fungicide regimens
furniture - The types of furniture present in the sampled room
gaseous_environment - Use of conditions with differing gaseous environments; should include the name of gaseous compound, amount administered, treatment duration, interval and total experimental duration; can include multiple gaseous environment regimens
gaseous_substances - Amount or concentration of substances such as hydrogen sulfide, carbon dioxide, methane, etc.; can include multiple substances
gastrointest_disord - History of gastrointestinal tract disorders; can include multiple disorders
gender_restroom - The gender type of the restroom
genetic_mod - Genetic modifications of the genome of an organism, which may occur naturally by spontaneous mutation, or be introduced by some experimental means, e.g. specification of a transgene or the gene knocked-out or details of transient transfection
geo_loc_name - The geographical origin of the sample as defined by the country or sea name followed by specific region name. Country or sea names should be chosen from the INSDC country list (http://insdc.org/country.html), or the GAZ ontology (v 1.512) (http://purl.bioontology.org/ontology/GAZ)
gestation_state - Specification of the gestation state
git url - Example: https://github.com/microbiomedata/mg_annotation/releases/tag/0.1
glucosidase_act - Measurement of glucosidase activity
gold_path_field - This is a grouping for any of the gold path fields
ecosystem - An ecosystem is a combination of a physical environment (abiotic factors) and all the organisms (biotic factors) that interact with this environment. Ecosystem is in position 1/5 in a GOLD path.
ecosystem_category - Ecosystem categories represent divisions within the ecosystem based on specific characteristics of the environment from where an organism or sample is isolated. Ecosystem category is in position 2/5 in a GOLD path.
ecosystem_subtype - Ecosystem subtypes represent further subdivision of Ecosystem types into more distinct subtypes. Ecosystem subtype is in position 4/5 in a GOLD path.
ecosystem_type - Ecosystem types represent things having common characteristics within the Ecosystem Category. These common characteristics based grouping is still broad but specific to the characteristics of a given environment. Ecosystem type is in position 3/5 in a GOLD path.
specific_ecosystem - Specific ecosystems represent specific features of the environment like aphotic zone in an ocean or gastric mucosa within a host digestive system. Specific ecosystem is in position 5/5 in a GOLD path.
gravidity - Whether or not subject is gravid, and if yes date due or date post-conception, specifying which is used
gravity - Information about treatment involving use of gravity factor to study various types of responses in presence, absence or modified levels of gravity; treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple treatments
growth_facil - Type of facility where the sampled plant was grown; controlled vocabulary: growth chamber, open top chamber, glasshouse, experimental garden, field. Alternatively use Crop Ontology (CO) terms, see http://www.cropontology.org/ontology/CO_715/Crop%20Research
growth_habit - Characteristic shape, appearance or growth form of a plant species
growth_hormone_regm - Information about treatment involving use of growth hormones; should include the name of growth hormone, amount administered, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple growth hormone regimens
gynecologic_disord - History of gynecological disorders; can include multiple disorders
hall_count - The total count of hallways and cooridors in the built structure
handidness - The handidness of the individual sampled
hc_produced - Main hydrocarbon type produced from resource (i.e. Oil, gas, condensate, etc). If ‘other’ is specified, please propose entry in ‘additional info’ field
hcr - Main Hydrocarbon Resource type. The term ‘Hydrocarbon Resource’ HCR defined as a natural environmental feature containing large amounts of hydrocarbons at high concentrations potentially suitable for commercial exploitation. This term should not be confused with the Hydrocarbon Occurrence term which also includes hydrocarbon-rich environments with currently limited commercial interest such as seeps, outcrops, gas hydrates etc. If ‘other’ is specified, please propose entry in ‘additional info’ field
hcr_fw_salinity - Original formation water salinity (prior to secondary recovery e.g. Waterflooding) expressed as TDS
hcr_geol_age - Geological age of hydrocarbon resource (Additional info: https://en.wikipedia.org/wiki/Period_(geology)). If ‘other’ is specified, please propose entry in ‘additional info’ field
hcr_pressure - Original pressure of the hydrocarbon resource
hcr_temp - Original temperature of the hydrocarbon resource
health_disease_stat - Health or disease status of specific host at time of collection
heat_cool_type - Methods of conditioning or heating a room or building
heat_deliv_loc - The location of heat delivery within the room
heat_system_deliv_meth - The method by which the heat is delivered through the system
heat_system_id - The heating system identifier
heavy_metals - Heavy metals present and concentrationsany drug used by subject and the frequency of usage; can include multiple heavy metals and concentrations
heavy_metals_meth - Reference or method used in determining heavy metals
height_carper_fiber - The average carpet fiber height in the indoor environment
herbicide_regm - Information about treatment involving use of herbicides; information about treatment involving use of growth hormones; should include the name of herbicide, amount administered, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple regimens
horizon - Specific layer in the land area which measures parallel to the soil surface and possesses physical characteristics which differ from the layers above and beneath
horizon_meth - Reference or method used in determining the horizon
host_age - Age of host at the time of sampling; relevant scale depends on species and study, e.g. Could be seconds for amoebae or centuries for trees
host_blood_press_diast - Resting diastolic blood pressure, measured as mm mercury
host_blood_press_syst - Resting systolic blood pressure, measured as mm mercury
host_body_habitat - Original body habitat where the sample was obtained from
host_body_mass_index - Body mass index, calculated as weight/(height)squared
host_body_product - Substance produced by the body, e.g. Stool, mucus, where the sample was obtained from. For foundational model of anatomy ontology (fma) or Uber-anatomy ontology (UBERON) terms, please see https://www.ebi.ac.uk/ols/ontologies/fma or https://www.ebi.ac.uk/ols/ontologies/uberon
host_body_site - Name of body site where the sample was obtained from, such as a specific organ or tissue (tongue, lung etc…). For foundational model of anatomy ontology (fma) (v 4.11.0) or Uber-anatomy ontology (UBERON) (v releases/2014-06-15) terms, please see http://purl.bioontology.org/ontology/FMA or http://purl.bioontology.org/ontology/UBERON
host_body_temp - Core body temperature of the host when sample was collected
host_color - The color of host
host_common_name - Common name of the host, e.g. Human
host_diet - Type of diet depending on the host, for animals omnivore, herbivore etc., for humans high-fat, meditteranean etc.; can include multiple diet types
host_disease_stat - List of diseases with which the host has been diagnosed; can include multiple diagnoses. The value of the field depends on host; for humans the terms should be chosen from do (disease ontology) at http://www.disease-ontology.org, other hosts are free text
host_dry_mass - Measurement of dry mass
host_family_relationship - Relationships to other hosts in the same study; can include multiple relationships
host_genotype - Observed genotype
host_growth_cond - Literature reference giving growth conditions of the host
host_height - The height of subject
host_hiv_stat - HIV status of subject, if yes HAART initiation status should also be indicated as [YES or NO]
host_infra_specific_name - Taxonomic information about the host below subspecies level
host_infra_specific_rank - Taxonomic rank information about the host below subspecies level, such as variety, form, rank etc.
host_last_meal - Content of last meal and time since feeding; can include multiple values
host_length - The length of subject
host_life_stage - Description of life stage of host
host_occupation - Most frequent job performed by subject
host_phenotype - Phenotype of human or other host. For phenotypic quality ontology (pato) (v 2018-03-27) terms, please see http://purl.bioontology.org/ontology/pato. For Human Phenotype Ontology (HP) (v 2018-06-13) please see http://purl.bioontology.org/ontology/HP
host_pred_appr - Tool or approach used for host prediction
host_pred_est_acc - For each tool or approach used for host prediction, estimated false discovery rates should be included, either computed de novo or from the literature
host_pulse - Resting pulse, measured as beats per minute
host_sex - Physical sex of the host
host_shape - Morphological shape of host
host_spec_range - The NCBI taxonomy identifier of the specific host if it is known
host_subject_id - A unique identifier by which each subject can be referred to, de-identified, e.g. #131
host_substrate - The growth substrate of the host
host_taxid - NCBI taxon id of the host, e.g. 9606
host_tot_mass - Total mass of the host at collection, the unit depends on host
host_wet_mass - Measurement of wet mass
hrt - Whether subject had hormone replacement theraphy, and if yes start date
humidity - Amount of water vapour in the air, at the time of sampling
humidity_regm - Information about treatment involving an exposure to varying degree of humidity; information about treatment involving use of growth hormones; should include amount of humidity administered, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple regimens
hysterectomy - Specification of whether hysterectomy was performed
ihmc_ethnicity - Ethnicity of the subject
ihmc_medication_code - Can include multiple medication codes
indoor_space - A distinguishable space within a structure, the purpose for which discrete areas of a building is used
indoor_surf - Type of indoor surface
indust_eff_percent - Percentage of industrial effluents received by wastewater treatment plant
inorg_particles - Concentration of particles such as sand, grit, metal particles, ceramics, etc.; can include multiple particles
inside_lux - The recorded value at sampling time (power density)
int_wall_cond - The physical condition of the wall at the time of sampling; photos or video preferred; use drawings to indicate location of damaged areas
investigation_type - Nucleic Acid Sequence Report is the root element of all MIGS/MIMS compliant reports as standardized by Genomic Standards Consortium. This field is either eukaryote,bacteria,virus,plasmid,organelle, metagenome,mimarks-survey, mimarks-specimen, metatranscriptome, single amplified genome, metagenome-assembled genome, or uncultivated viral genome
isol_growth_condt - Publication reference in the form of pubmed ID (pmid), digital object identifier (doi) or url for isolation and growth condition specifications of the organism/material
iw_bt_date_well - Injection water breakthrough date per well following a secondary and/or tertiary recovery
iwf - Proportion of the produced fluids derived from injected water at the time of sampling. (e.g. 87%)
kidney_disord - History of kidney disorders; can include multiple disorders
last_clean - The last time the floor was cleaned (swept, mopped, vacuumed)
lat_lon - The geographical origin of the sample as defined by latitude and longitude. The values should be reported in decimal degrees and in WGS84 system
biosample➞lat_lon - This is currently a required field but it’s not clear if this should be required for human hosts
lib_layout - Specify whether to expect single, paired, or other configuration of reads
lib_reads_seqd - Total number of clones sequenced from the library
lib_screen - Specific enrichment or screening methods applied before and/or after creating libraries
lib_size - Total number of clones in the library prepared for the project
lib_vector - Cloning vector type(s) used in construction of libraries
light_intensity - Measurement of light intensity
light_regm - Information about treatment(s) involving exposure to light, including both light intensity and quality.
light_type - Application of light to achieve some practical or aesthetic effect. Lighting includes the use of both artificial light sources such as lamps and light fixtures, as well as natural illumination by capturing daylight. Can also include absence of light
link_addit_analys - Link to additional analysis results performed on the sample
link_class_info - Link to digitized soil maps or other soil classification information
link_climate_info - Link to climate resource
lithology - Hydrocarbon resource main lithology (Additional information: http://petrowiki.org/Lithology_and_rock_type_determination). If ‘other’ is specified, please propose entry in ‘additional info’ field
liver_disord - History of liver disorders; can include multiple disorders
local_class - Soil classification based on local soil classification system
local_class_meth - Reference or method used in determining the local soil classification
mag_cov_software - Tool(s) used to determine the genome coverage if coverage is used as a binning parameter in the extraction of genomes from metagenomic datasets
magnesium - Concentration of magnesium in the sample
maternal_health_stat - Specification of the maternal health status
max_occup - The maximum amount of people allowed in the indoor environment
md5 checksum - MD5 checksum of file (pre-compressed)
mean_frict_vel - Measurement of mean friction velocity
mean_peak_frict_vel - Measurement of mean peak friction velocity
mech_struc - mechanical structure: a moving structure
mechanical_damage - Information about any mechanical damage exerted on the plant; can include multiple damages and sites
medic_hist_perform - Whether full medical history was collected
menarche - Date of most recent menstruation
menopause - Date of onset of menopause
methane - Methane (gas) amount or concentration at the time of sampling
microbial_biomass - The part of the organic matter in the soil that constitutes living microorganisms smaller than 5-10 micrometer. If you keep this, you would need to have correction factors used for conversion to the final units
microbial_biomass_meth - Reference or method used in determining microbial biomass
mid - Molecular barcodes, called Multiplex Identifiers (MIDs), that are used to specifically tag unique samples in a sequencing run. Sequence should be reported in uppercase letters
mineral_nutr_regm - Information about treatment involving the use of mineral supplements; should include the name of mineral nutrient, amount administered, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple mineral nutrient regimens
misc_param - Any other measurement performed or parameter collected, that is not listed here
n_alkanes - Concentration of n-alkanes; can include multiple n-alkanes
nitrate - Concentration of nitrate in the sample
nitrite - Concentration of nitrite in the sample
nitro - Concentration of nitrogen (total)
non_mineral_nutr_regm - Information about treatment involving the exposure of plant to non-mineral nutrient such as oxygen, hydrogen or carbon; should include the name of non-mineral nutrient, amount administered, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple non-mineral nutrient regimens
nose_mouth_teeth_throat_disord - History of nose/mouth/teeth/throat disorders; can include multiple disorders
nose_throat_disord - History of nose-throat disorders; can include multiple disorders
nucl_acid_amp - A link to a literature reference, electronic resource or a standard operating procedure (SOP), that describes the enzymatic amplification (PCR, TMA, NASBA) of specific nucleic acids
nucl_acid_ext - A link to a literature reference, electronic resource or a standard operating procedure (SOP), that describes the material separation to recover the nucleic acid fraction from a sample
num_replicons - Reports the number of replicons in a nuclear genome of eukaryotes, in the genome of a bacterium or archaea or the number of segments in a segmented virus. Always applied to the haploid chromosome count of a eukaryote
number_contig - Total number of contigs in the cleaned/submitted assembly that makes up a given genome, SAG, MAG, or UViG
number_pets - The number of pets residing in the sampled space
number_plants - The number of plant(s) in the sampling space
number_resident - The number of individuals currently occupying in the sampling location
occup_density_samp - Average number of occupants at time of sampling per square footage
occup_document - The type of documentation of occupancy
occup_samp - Number of occupants present at time of sample within the given space
org_carb - Concentration of organic carbon
org_matter - Concentration of organic matter
org_nitro - Concentration of organic nitrogen
org_particles - Concentration of particles such as faeces, hairs, food, vomit, paper fibers, plant material, humus, etc.
organism_count - Total cell count of any organism (or group of organisms) per gram, volume or area of sample, should include name of organism followed by count. The method that was used for the enumeration (e.g. qPCR, atp, mpn, etc.) Should also be provided. (example: total prokaryotes; 3.5e7 cells per ml; qpcr)
organism_count_qpcr_info - If qpcr was used for the cell count, the target gene name, the primer sequence and the cycling conditions should also be provided. (Example: 16S rrna; FWD:ACGTAGCTATGACGT REV:GTGCTAGTCGAGTAC; initial denaturation:90C_5min; denaturation:90C_2min; annealing:52C_30 sec; elongation:72C_30 sec; 90 C for 1 min; final elongation:72C_5min; 30 cycles)
owc_tvdss - Depth of the original oil water contact (OWC) zone (average) (m TVDSS)
oxy_stat_samp - Oxygenation status of sample
oxygen - Oxygen (gas) amount or concentration at the time of sampling
part_org_carb - Concentration of particulate organic carbon
part_org_nitro - Concentration of particulate organic nitrogen
particle_class - Particles are classified, based on their size, into six general categories:clay, silt, sand, gravel, cobbles, and boulders; should include amount of particle preceded by the name of the particle type; can include multiple values
pathogenicity - To what is the entity pathogenic
pcr_cond - Description of reaction conditions and components of PCR in the form of ‘initial denaturation:94degC_1.5min; annealing=…’
pcr_primers - PCR primers that were used to amplify the sequence of the targeted gene, locus or subfragment. This field should contain all the primers used for a single PCR reaction if multiple forward or reverse primers are present in a single PCR reaction. The primer sequence should be reported in uppercase letters
permeability - Measure of the ability of a hydrocarbon resource to allow fluids to pass through it. (Additional information: https://en.wikipedia.org/wiki/Permeability_(earth_sciences))
perturbation - Type of perturbation, e.g. chemical administration, physical disturbance, etc., coupled with perturbation regimen including how many times the perturbation was repeated, how long each perturbation lasted, and the start and end time of the entire perturbation period; can include multiple perturbation types
pesticide_regm - Information about treatment involving use of insecticides; should include the name of pesticide, amount administered, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple pesticide regimens
pet_farm_animal - Specification of presence of pets or farm animals in the environment of subject, if yes the animals should be specified; can include multiple animals present
petroleum_hydrocarb - Concentration of petroleum hydrocarbon
ph - Ph measurement of the sample, or liquid portion of sample, or aqueous phase of the fluid
ph_meth - Reference or method used in determining ph
ph_regm - Information about treatment involving exposure of plants to varying levels of ph of the growth media, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple regimen
phaeopigments - Concentration of phaeopigments; can include multiple phaeopigments
phosphate - Concentration of phosphate
phosplipid_fatt_acid - Concentration of phospholipid fatty acids; can include multiple values
photon_flux - Measurement of photon flux
plant_growth_med - Specification of the media for growing the plants or tissue cultured samples, e.g. soil, aeroponic, hydroponic, in vitro solid culture medium, in vitro liquid culture medium. Recommended value is a specific value from EO:plant growth medium (follow this link for terms http://purl.obolibrary.org/obo/EO_0007147) or other controlled vocabulary
plant_product - Substance produced by the plant, where the sample was obtained from
plant_sex - Sex of the reproductive parts on the whole plant, e.g. pistillate, staminate, monoecieous, hermaphrodite.
plant_struc - Name of plant structure the sample was obtained from; for Plant Ontology (PO) (v releases/2017-12-14) terms, see http://purl.bioontology.org/ontology/PO, e.g. petiole epidermis (PO_0000051). If an individual flower is sampled, the sex of it can be recorded here.
ploidy - The ploidy level of the genome (e.g. allopolyploid, haploid, diploid, triploid, tetraploid). It has implications for the downstream study of duplicated gene and regions of the genomes (and perhaps for difficulties in assembly). For terms, please select terms listed under class ploidy (PATO:001374) of Phenotypic Quality Ontology (PATO), and for a browser of PATO (v 2018-03-27) please refer to http://purl.bioontology.org/ontology/PATO
pollutants - Pollutant types and, amount or concentrations measured at the time of sampling; can report multiple pollutants by entering numeric values preceded by name of pollutant
pool_dna_extracts - Indicate whether multiple DNA extractions were mixed. If the answer yes, the number of extracts that were pooled should be given
porosity - Porosity of deposited sediment is volume of voids divided by the total volume of sample
potassium - Concentration of potassium in the sample
pour_point - Temperature at which a liquid becomes semi solid and loses its flow characteristics. In crude oil a highB pour pointB is generally associated with a high paraffin content, typically found in crude deriving from a larger proportion of plant material. (soure: https://en.wikipedia.org/wiki/pour_point)
pre_treatment - The process of pre-treatment removes materials that can be easily collected from the raw wastewater
pred_genome_struc - Expected structure of the viral genome
pred_genome_type - Type of genome predicted for the UViG
pregnancy - Date due of pregnancy
pres_animal - The number and type of animals present in the sampling space
pressure - Pressure to which the sample is subject to, in atmospheres
previous_land_use - Previous land use and dates
previous_land_use_meth - Reference or method used in determining previous land use and dates
primary_prod - Measurement of primary production, generally measured as isotope uptake
primary_treatment - The process to produce both a generally homogeneous liquid capable of being treated biologically and a sludge that can be separately treated or processed
principal investigator - Principal Investigator who led the study and/or generated the dataset.
prod_rate - Oil and/or gas production rates per well (e.g. 524 m3 / day)
prod_start_date - Date of field’s first production
profile_position - Cross-sectional position in the hillslope where sample was collected.sample area position in relation to surrounding areas
project_name - Name of the project within which the sequencing was organized
propagation - This field is specific to different taxa. For phages: lytic/lysogenic, for plasmids: incompatibility group, for eukaryotes: sexual/asexual (Note: there is the strong opinion to name phage propagation obligately lytic or temperate, therefore we also give this choice
pulmonary_disord - History of pulmonary disorders; can include multiple disorders
quad_pos - The quadrant position of the sampling room within the building
radiation_regm - Information about treatment involving exposure of plant or a plant part to a particular radiation regimen; should include the radiation type, amount or intensity administered, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple radiation regimens
rainfall_regm - Information about treatment involving an exposure to a given amount of rainfall, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple regimens
reactor_type - Anaerobic digesters can be designed and engineered to operate using a number of different process configurations, as batch or continuous, mesophilic, high solid or low solid, and single stage or multistage
reassembly_bin - Has an assembly been performed on a genome bin extracted from a metagenomic assembly?
redox_potential - Redox potential, measured relative to a hydrogen cell, indicating oxidation or reduction potential
ref_biomaterial - Primary publication if isolated before genome publication; otherwise, primary genome report
ref_db - List of database(s) used for ORF annotation, along with version number and reference to website or publication
rel_air_humidity - Partial vapor and air pressure, density of the vapor and air, or by the actual mass of the vapor and air
rel_humidity_out - The recorded outside relative humidity value at the time of sampling
rel_samp_loc - The sampling location within the train car
rel_to_oxygen - Is this organism an aerobe, anaerobe? Please note that aerobic and anaerobic are valid descriptors for microbial environments
reservoir - Name of the reservoir (e.g. Carapebus)
resins_pc - Saturate, Aromatic, Resin and AsphalteneB (SARA) is an analysis method that dividesB crude oilB components according to their polarizability and polarity. There are three main methods to obtain SARA results. The most popular one is known as the Iatroscan TLC-FID and is referred to as IP-143 (source: https://en.wikipedia.org/wiki/Saturate,_aromatic,_resin_and_asphaltene)
resp_part_matter - Concentration of substances that remain suspended in the air, and comprise mixtures of organic and inorganic substances (PM10 and PM2.5); can report multiple PM’s by entering numeric values preceded by name of PM
room_air_exch_rate - The rate at which outside air replaces indoor air in a given space
room_architec_element - The unique details and component parts that, together, form the architecture of a distinguisahable space within a built structure
room_condt - The condition of the room at the time of sampling
room_connected - List of rooms connected to the sampling room by a doorway
room_count - The total count of rooms in the built structure including all room types
room_dim - The length, width and height of sampling room
room_door_dist - Distance between doors (meters) in the hallway between the sampling room and adjacent rooms
room_door_share - List of room(s) (room number, room name) sharing a door with the sampling room
room_hallway - List of room(s) (room number, room name) located in the same hallway as sampling room
room_loc - The position of the room within the building
room_moist_damage_hist - The history of moisture damage or mold in the past 12 months. Number of events of moisture damage or mold observed
room_net_area - The net floor area of sampling room. Net area excludes wall thicknesses
room_occup - Count of room occupancy at time of sampling
room_samp_pos - The horizontal sampling position in the room relative to architectural elements
room_type - The main purpose or activity of the sampling room. A room is any distinguishable space within a structure
room_vol - Volume of sampling room
room_wall_share - List of room(s) (room number, room name) sharing a wall with the sampling room
room_window_count - Number of windows in the room
root_cond - Relevant rooting conditions such as field plot size, sowing density, container dimensions, number of plants per container.
root_med_carbon - Source of organic carbon in the culture rooting medium; e.g. sucrose.
root_med_macronutr - Measurement of the culture rooting medium macronutrients (N,P, K, Ca, Mg, S); e.g. KH2PO4 (170B mg/L).
root_med_micronutr - Measurement of the culture rooting medium micronutrients (Fe, Mn, Zn, B, Cu, Mo); e.g. H3BO3 (6.2B mg/L).
root_med_ph - pH measurement of the culture rooting medium; e.g. 5.5.
root_med_regl - Growth regulators in the culture rooting medium such as cytokinins, auxins, gybberellins, abscisic acid; e.g. 0.5B mg/L NAA.
root_med_solid - Specification of the solidifying agent in the culture rooting medium; e.g. agar.
root_med_suppl - Organic supplements of the culture rooting medium, such as vitamins, amino acids, organic acids, antibiotics activated charcoal; e.g. nicotinic acid (0.5B mg/L).
salinity - Salinity is the total concentration of all dissolved salts in a water sample. While salinity can be measured by a complete chemical analysis, this method is difficult and time consuming. More often, it is instead derived from the conductivity measurement. This is known as practical salinity. These derivations compare the specific conductance of the sample to a salinity standard such as seawater
salinity_meth - Reference or method used in determining salinity
salt_regm - Information about treatment involving use of salts as supplement to liquid and soil growth media; should include the name of salt, amount administered, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple salt regimens
samp_capt_status - Reason for the sample
samp_collect_device - The method or device employed for collecting the sample
samp_collection_point - Sampling point on the asset were sample was collected (e.g. Wellhead, storage tank, separator, etc). If ‘other’ is specified, please propose entry in ‘additional info’ field
samp_dis_stage - Stage of the disease at the time of sample collection, e.g. inoculation, penetration, infection, growth and reproduction, dissemination of pathogen.
samp_floor - The floor of the building, where the sampling room is located
samp_loc_corr_rate - Metal corrosion rate is the speed of metal deterioration due to environmental conditions. As environmental conditions change corrosion rates change accordingly. Therefore, long term corrosion rates are generally more informative than short term rates and for that reason they are preferred during reporting. In the case of suspected MIC, corrosion rate measurements at the time of sampling might provide insights into the involvement of certain microbial community members in MIC as well as potential microbial interplays
samp_mat_process - Any processing applied to the sample during or after retrieving the sample from environment. This field accepts OBI, for a browser of OBI (v 2018-02-12) terms please see http://purl.bioontology.org/ontology/OBI
samp_md - In non deviated well, measured depth is equal to the true vertical depth, TVD (TVD=TVDSS plus the reference or datum it refers to). In deviated wells, the MD is the length of trajectory of the borehole measured from the same reference or datum. Common datums used are ground level (GL), drilling rig floor (DF), rotary table (RT), kelly bushing (KB) and mean sea level (MSL). If ‘other’ is specified, please propose entry in ‘additional info’ field
samp_preserv - Preservative added to the sample (e.g. Rnalater, alcohol, formaldehyde, etc.). Where appropriate include volume added (e.g. Rnalater; 2 ml)
samp_room_id - Sampling room number. This ID should be consistent with the designations on the building floor plans
samp_salinity - Salinity is the total concentration of all dissolved salts in a liquid or solid (in the form of an extract obtained by centrifugation) sample. While salinity can be measured by a complete chemical analysis, this method is difficult and time consuming. More often, it is instead derived from the conductivity measurement. This is known as practical salinity. These derivations compare the specific conductance of the sample to a salinity standard such as seawater
samp_size - Amount or size of sample (volume, mass or area) that was collected
samp_sort_meth - Method by which samples are sorted; open face filter collecting total suspended particles, prefilter to remove particles larger than X micrometers in diameter, where common values of X would be 10 and 2.5 full size sorting in a cascade impactor.
samp_store_dur - Duration for which the sample was stored
samp_store_loc - Location at which sample was stored, usually name of a specific freezer/room
samp_store_temp - Temperature at which sample was stored, e.g. -80 degree Celsius
samp_subtype - Name of sample sub-type. For example if ‘sample type’ is ‘Produced Water’ then subtype could be ‘Oil Phase’ or ‘Water Phase’. If ‘other’ is specified, please propose entry in ‘additional info’ field
samp_time_out - The recent and long term history of outside sampling
samp_transport_cond - Sample transport duration (in days or hrs) and temperature the sample was exposed to (e.g. 5.5 days; 20 B0C)
samp_tvdss - Depth of the sample i.e. The vertical distance between the sea level and the sampled position in the subsurface. Depth can be reported as an interval for subsurface samples e.g. 1325.75-1362.25 m
samp_type - Type of material (i.e. sample) collected. Includes types like core, rock trimmings, drill cuttings, piping section, coupon, pigging debris, solid deposit, produced fluid, produced water, injected water, swabs, etc. If ‘other’ is specified, please propose entry in ‘additional info’ field
samp_vol_we_dna_ext - Volume (ml), weight (g) of processed sample, or surface area swabbed from sample for DNA extraction
samp_weather - The weather on the sampling day
samp_well_name - Name of the well (e.g. BXA1123) where sample was taken
saturates_pc - Saturate, Aromatic, Resin and AsphalteneB (SARA) is an analysis method that dividesB crude oilB components according to their polarizability and polarity. There are three main methods to obtain SARA results. The most popular one is known as the Iatroscan TLC-FID and is referred to as IP-143 (source: https://en.wikipedia.org/wiki/Saturate,_aromatic,_resin_and_asphaltene)
season - The season when sampling occurred
season_environment - Treatment involving an exposure to a particular season (e.g. Winter, summer, rabi, rainy etc.), treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment
season_precpt - The average of all seasonal precipitation values known, or an estimated equivalent value derived by such methods as regional indexes or Isohyetal maps.
season_temp - Mean seasonal temperature
season_use - The seasons the space is occupied
secondary_treatment - The process for substantially degrading the biological content of the sewage
sediment_type - Information about the sediment type based on major constituents
seq_meth - Sequencing method used; e.g. Sanger, pyrosequencing, ABI-solid
seq_quality_check - Indicate if the sequence has been called by automatic systems (none) or undergone a manual editing procedure (e.g. by inspecting the raw data or chromatograms). Applied only for sequences that are not submitted to SRA,ENA or DRA
sewage_type - Type of wastewater treatment plant as municipial or industrial
sexual_act - Current sexual partner and frequency of sex
shading_device_cond - The physical condition of the shading device at the time of sampling
shading_device_loc - The location of the shading device in relation to the built structure
shading_device_mat - The material the shading device is composed of
shading_device_type - The type of shading device
shading_device_water_mold - Signs of the presence of mold or mildew on the shading device
sieving - Collection design of pooled samples and/or sieve size and amount of sample sieved
silicate - Concentration of silicate
sim_search_meth - Tool used to compare ORFs with database, along with version and cutoffs used
single_cell_lysis_appr - Method used to free DNA from interior of the cell(s) or particle(s)
single_cell_lysis_prot - Name of the kit or standard protocol used for cell(s) or particle(s) lysis
size_frac - Filtering pore size used in sample preparation
size_frac_low - Refers to the mesh/pore size used to pre-filter/pre-sort the sample. Materials larger than the size threshold are excluded from the sample
size_frac_up - Refers to the mesh/pore size used to retain the sample. Materials smaller than the size threshold are excluded from the sample
slope_aspect - The direction a slope faces. While looking down a slope use a compass to record the direction you are facing (direction or degrees); e.g., nw or 315 degrees. This measure provides an indication of sun and wind exposure that will influence soil temperature and evapotranspiration.
slope_gradient - Commonly called ‘slope’. The angle between ground surface and a horizontal line (in percent). This is the direction that overland water would flow. This measure is usually taken with a hand level meter or clinometer
sludge_retent_time - The time activated sludge remains in reactor
smoker - Specification of smoking status
sodium - Sodium concentration in the sample
soil_type - Soil series name or other lower-level classification
soil_type_meth - Reference or method used in determining soil series name or other lower-level classification
solar_irradiance - The amount of solar energy that arrives at a specific area of a surface during a specific time interval
soluble_inorg_mat - Concentration of substances such as ammonia, road-salt, sea-salt, cyanide, hydrogen sulfide, thiocyanates, thiosulfates, etc.
soluble_org_mat - Concentration of substances such as urea, fruit sugars, soluble proteins, drugs, pharmaceuticals, etc.
soluble_react_phosp - Concentration of soluble reactive phosphorus
sop - Standard operating procedures used in assembly and/or annotation of genomes, metagenomes or environmental sequences
sort_tech - Method used to sort/isolate cells or particles of interest
source_mat_id - A unique identifier assigned to a material sample (as defined by http://rs.tdwg.org/dwc/terms/materialSampleID, and as opposed to a particular digital record of a material sample) used for extracting nucleic acids, and subsequent sequencing. The identifier can refer either to the original material collected or to any derived sub-samples. The INSDC qualifiers /specimen_voucher, /bio_material, or /culture_collection may or may not share the same value as the source_mat_id field. For instance, the /specimen_voucher qualifier and source_mat_id may both contain ‘UAM:Herps:14’ , referring to both the specimen voucher and sampled tissue with the same identifier. However, the /culture_collection qualifier may refer to a value from an initial culture (e.g. ATCC:11775) while source_mat_id would refer to an identifier from some derived culture from which the nucleic acids were extracted (e.g. xatc123 or ark:/2154/R2).
source_uvig - Type of dataset from which the UViG was obtained
space_typ_state - Customary or normal state of the space
special_diet - Specification of special diet; can include multiple special diets
specific - The building specifications. If design is chosen, indicate phase: conceptual, schematic, design development, construction documents
specific_host - If there is a host involved, please provide its taxid (or environmental if not actually isolated from the dead or alive host - i.e. a pathogen could be isolated from a swipe of a bench etc) and report whether it is a laboratory or natural host)
specific_humidity - The mass of water vapour in a unit mass of moist air, usually expressed as grams of vapour per kilogram of air, or, in air conditioning, as grains per pound.
sr_dep_env - Source rock depositional environment (https://en.wikipedia.org/wiki/Source_rock). If ‘other’ is specified, please propose entry in ‘additional info’ field
sr_geol_age - Geological age of source rock (Additional info: https://en.wikipedia.org/wiki/Period_(geology)). If ‘other’ is specified, please propose entry in ‘additional info’ field
sr_kerog_type - Origin of kerogen. Type I: Algal (aquatic), Type II: planktonic and soft plant material (aquatic or terrestrial), Type III: terrestrial woody/ fibrous plant material (terrestrial), Type IV: oxidized recycled woody debris (terrestrial) (additional information: https://en.wikipedia.org/wiki/Kerogen). If ‘other’ is specified, please propose entry in ‘additional info’ field
sr_lithology - Lithology of source rock (https://en.wikipedia.org/wiki/Source_rock). If ‘other’ is specified, please propose entry in ‘additional info’ field
standing_water_regm - Treatment involving an exposure to standing water during a plant’s life span, types can be flood water or standing water, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple regimens
store_cond - Explain how and for how long the soil sample was stored before DNA extraction
study_complt_stat - Specification of study completion status, if no the reason should be specified
submitted_to_insdc - Depending on the study (large-scale e.g. done with next generation sequencing technology, or small-scale) sequences have to be submitted to SRA (Sequence Read Archive), DRA (DDBJ Read Archive) or via the classical Webin/Sequin systems to Genbank, ENA and DDBJ. Although this field is mandatory, it is meant as a self-test field, therefore it is not necessary to include this field in contextual data submitted to databases
subspecf_gen_lin - This should provide further information about the genetic distinctness of the sequenced organism by recording additional information e.g. serovar, serotype, biotype, ecotype, or any relevant genetic typing schemes like Group I plasmid. It can also contain alternative taxonomic information. It should contain both the lineage name, and the lineage rank, i.e. biovar:abc123
substructure_type - The substructure or under building is that largely hidden section of the building which is built off the foundations to the ground floor level
sulfate - Concentration of sulfate in the sample
sulfate_fw - Original sulfate concentration in the hydrocarbon resource
sulfide - Concentration of sulfide in the sample
surf_air_cont - Contaminant identified on surface
surf_humidity - Surfaces: water activity as a function of air and material moisture
surf_material - Surface materials at the point of sampling
surf_moisture - Water held on a surface
surf_moisture_ph - ph measurement of surface
surf_temp - Temperature of the surface at the time of sampling
suspend_part_matter - Concentration of suspended particulate matter
suspend_solids - Concentration of substances including a wide variety of material, such as silt, decaying plant and animal matter; can include multiple substances
tan - Total Acid NumberB (TAN) is a measurement of acidity that is determined by the amount ofB potassium hydroxideB in milligrams that is needed to neutralize the acids in one gram of oil.B It is an important quality measurement ofB crude oil. (source: https://en.wikipedia.org/wiki/Total_acid_number)
target_gene - Targeted gene or locus name for marker gene studies
target_subfragment - Name of subfragment of a gene or locus. Important to e.g. identify special regions on marker genes like V6 on 16S rRNA
tax_class - Method used for taxonomic classification, along with reference database used, classification rank, and thresholds used to classify new genomes
tax_ident - The phylogenetic marker(s) used to assign an organism name to the SAG or MAG
temp - Temperature of the sample at the time of sampling
temp_out - The recorded temperature value at sampling time outside
tertiary_treatment - The process providing a final treatment stage to raise the effluent quality before it is discharged to the receiving environment
texture - The relative proportion of different grain sizes of mineral particles in a soil, as described using a standard system; express as % sand (50 um to 2 mm), silt (2 um to 50 um), and clay (<2 um) with textural name (e.g., silty clay loam) optional.
texture_meth - Reference or method used in determining soil texture
tidal_stage - Stage of tide
tillage - Note method(s) used for tilling
time_last_toothbrush - Specification of the time since last toothbrushing
time_since_last_wash - Specification of the time since last wash
tiss_cult_growth_med - Description of plant tissue culture growth media used
toluene - Concentration of toluene in the sample
tot_carb - Total carbon content
tot_depth_water_col - Measurement of total depth of water column
tot_diss_nitro - Total dissolved nitrogen concentration, reported as nitrogen, measured by: total dissolved nitrogen = NH4 + NO3NO2 + dissolved organic nitrogen
tot_inorg_nitro - Total inorganic nitrogen content
tot_iron - Concentration of total iron in the sample
tot_nitro - Total nitrogen concentration of water samples, calculated by: total nitrogen = total dissolved nitrogen + particulate nitrogen. Can also be measured without filtering, reported as nitrogen
tot_nitro_content - Total nitrogen content of the sample
tot_nitro_content_meth - Reference or method used in determining the total nitrogen
tot_org_c_meth - Reference or method used in determining total organic carbon
tot_org_carb - Definition for soil: total organic carbon content of the soil, definition otherwise: total organic carbon content
tot_part_carb - Total particulate carbon content
tot_phosp - Total phosphorus concentration in the sample, calculated by: total phosphorus = total dissolved phosphorus + particulate phosphorus
tot_phosphate - Total amount or concentration of phosphate
tot_sulfur - Concentration of total sulfur in the sample
train_line - The subway line name
train_stat_loc - The train station collection location
train_stop_loc - The train stop collection location
travel_out_six_month - Specification of the countries travelled in the last six months; can include multiple travels
trna_ext_software - Tools used for tRNA identification
trnas - The total number of tRNAs identified from the SAG or MAG
trophic_level - Trophic levels are the feeding position in a food chain. Microbes can be a range of producers (e.g. chemolithotroph)
turbidity - Measure of the amount of cloudiness or haziness in water caused by individual particles
tvdss_of_hcr_pressure - True vertical depth subsea (TVDSS) of the hydrocarbon resource where the original pressure was measured (e.g. 1578 m )
tvdss_of_hcr_temp - True vertical depth subsea (TVDSS) of the hydrocarbon resource where the original temperature was measured (e.g. 1345 m)
twin_sibling - Specification of twin sibling presence
typ_occup_density - Customary or normal density of occupants
urine_collect_meth - Specification of urine collection method
urogenit_disord - History of urogenital disorders, can include multiple disorders
urogenit_tract_disor - History of urogenitaltract disorders; can include multiple disorders
ventilation_rate - Ventilation rate of the system in the sampled premises
ventilation_type - Ventilation system used in the sampled premises
vfa - Concentration of Volatile Fatty Acids in the sample
vfa_fw - Original volatile fatty acid concentration in the hydrocarbon resource
vir_ident_software - Tool(s) used for the identification of UViG as a viral genome, software or protocol name including version number, parameters, and cutoffs used
virus_enrich_appr - List of approaches used to enrich the sample for viruses, if any
vis_media - The building visual media
viscosity - A measure of oil’s resistanceB to gradual deformation byB shear stressB orB tensile stress (e.g. 3.5 cp; 100 B0C)
volatile_org_comp - Concentration of carbon-based chemicals that easily evaporate at room temperature; can report multiple volatile organic compounds by entering numeric values preceded by name of compound
votu_class_appr - Cutoffs and approach used when clustering new UViGs in Rspecies-levelS vOTUs. Note that results from standard 95% ANI / 85% AF clustering should be provided alongside vOTUS defined from another set of thresholds, even if the latter are the ones primarily used during the analysis
votu_db - Reference database (i.e. sequences not generated as part of the current study) used to cluster new genomes in ‘species-level’ vOTUs, if any
votu_seq_comp_appr - Tool and thresholds used to compare sequences when computing ‘species-level’ vOTUs
wall_area - The total area of the sampled room’s walls
wall_const_type - The building class of the wall defined by the composition of the building elements and fire-resistance rating.
wall_finish_mat - The material utilized to finish the outer most layer of the wall
wall_height - The average height of the walls in the sampled room
wall_loc - The relative location of the wall within the room
wall_surf_treatment - The surface treatment of interior wall
wall_texture - The feel, appearance, or consistency of a wall surface
wall_thermal_mass - The ability of the wall to provide inertia against temperature fluctuations. Generally this means concrete or concrete block that is either exposed or covered only with paint
wall_water_mold - Signs of the presence of mold or mildew on a wall
wastewater_type - The origin of wastewater such as human waste, rainfall, storm drains, etc.
water_content - Water content measurement
water_content_soil_meth - Reference or method used in determining the water content of soil
water_current - Measurement of magnitude and direction of flow within a fluid
water_cut - Current amount of water (%) in a produced fluid stream; or the average of the combined streams
water_feat_size - The size of the water feature
water_feat_type - The type of water feature present within the building being sampled
water_production_rate - Water production rates per well (e.g. 987 m3 / day)
water_temp_regm - Information about treatment involving an exposure to water with varying degree of temperature, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple regimens
watering_regm - Information about treatment involving an exposure to watering frequencies, treatment regimen including how many times the treatment was repeated, how long each treatment lasted, and the start and end time of the entire treatment; can include multiple regimens
weekday - The day of the week when sampling occurred
weight_loss_3_month - Specification of weight loss in the last three months, if yes should be further specified to include amount of weight loss
wga_amp_appr - Method used to amplify genomic DNA in preparation for sequencing
wga_amp_kit - Kit used to amplify genomic DNA in preparation for sequencing
win - A unique identifier of a well or wellbore. This is part of the Global Framework for Well Identification initiative which is compiled by the Professional Petroleum Data Management Association (PPDM) in an effort to improve well identification systems. (Supporting information: https://ppdm.org/ and http://dl.ppdm.org/dl/690)
wind_direction - Wind direction is the direction from which a wind originates
wind_speed - Speed of wind measured at the time of sampling
window_cond - The physical condition of the window at the time of sampling
window_cover - The type of window covering
window_horiz_pos - The horizontal position of the window on the wall
window_loc - The relative location of the window within the room
window_mat - The type of material used to finish a window
window_open_freq - The number of times windows are opened per week
window_size - The window’s length and width
window_status - Defines whether the windows were open or closed during environmental testing
window_type - The type of windows
window_vert_pos - The vertical position of the window on the wall
window_water_mold - Signs of the presence of mold or mildew on the window.
xylene - Concentration of xylene in the sample
best protein - the specific protein identifier most correctly associated with the peptide sequence
protein quantification➞best protein - the specific protein identifier most correctly grouped to its associated peptide sequences
biosample set - This property links a database object to the set of samples within it.
chemical formula - A generic grouping for miolecular formulae and empirican formulae
compression type - If provided, specifies the compression type
data object set - This property links a database object to the set of data objects within it.
data object type - The type of file represented by the data object.
date created - TODO
description - a human-readable description of a thing
direction - One of l->r, r->l, bidirectional, neutral
ecosystem_path_id - A unique id representing the GOLD classifers associated with a sample.
email - An email address for an entity such as a person. This should be the primarly email address used.
encodes - The gene product encoded by this feature. Typically this is used for a CDS feature or gene feature which will encode a protein. It can also be used by a nc transcript ot gene feature that encoded a ncRNA
ess dive datasets - List of ESS-DIVE dataset DOIs
etl software version - TODO
feature type - TODO: Yuri to write
functional annotation set - This property links a database object to the set of all functional annotations
genome feature set - This property links a database object to the set of all features
gff coordinate - A positive 1-based integer coordinate indicating start or end
end - The end of the feature in positive 1-based integer coordinates
genome feature➞end - The end of the feature in positive 1-based integer coordinates
genome feature➞start - The start of the feature in positive 1-based integer coordinates
start - The start of the feature in positive 1-based integer coordinates
has boolean value - Links a quantity value to a boolean
has calibration - TODO: Yuri to fill in
nom analysis activity➞has calibration - A reference to a file that holds calibration information.
has credit associations - This slot links a study to a credit association. The credit association will be linked to a person value and to a CRediT Contributor Roles term. Overall semantics: person should get credit X for their participation in the study
has input - An input to a process.
has numeric value - Links a quantity value to a number
has maximum numeric value - The maximum value part, expressed as number, of the quantity value when the value covers a range.
has minimum numeric value - The minimum value part, expressed as number, of the quantity value when the value covers a range.
quantity value➞has numeric value - The number part of the quantity
has output - An output biosample to a processing step
has raw value - The value that was specified for an annotation in raw form, i.e. a string. E.g. “2 cm” or “2-4 cm”
geolocation value➞has raw value - The raw value for a geolocation should follow {lat} {long}
person value➞has raw value - The full name of the Investgator in format FIRST LAST.
quantity value➞has raw value - Unnormalized atomic string representation, should in syntax {number} {unit}
has unit - Links a quantity value to a unit
quantity value➞has unit - The unit of the quantity
has_part - A pathway can be broken down to a series of reaction step
highest similarity score - TODO: Yuri to fill in
id - A unique identifier for a thing. Must be either a CURIE shorthand for a URI or a complete URI
person➞id - Should be an ORCID. Specify in CURIE format. E.g ORCID:0000-1111-…
input_read_bases - TODO
instrument_name - The name of the instrument that was used for processing the sample.
is fully characterized - False if includes R-groups
keywords - A list of keywords that used to associate the entity. Keywords SHOULD come from controlled vocabularies, including MESH, ENVO.
language - Should use ISO 639-1 code e.g. “en”, “fr”
latitude - latitude
longitude - longitude
mags activity set - This property links a database object to the set of MAGs analysis activities.
metabolite quantified - the specific metabolite identifier
metabolomics analysis activity set - This property links a database object to the set of metabolomics analysis activities.
metagenome annotation activity set - This property links a database object to the set of metagenome annotation activities.
-
asm_score - A score for comparing metagenomic assembly quality from same sample.
contig_bp - Total size in bp of all contigs.
contigs - The sum of the (length*log(length)) of all contigs, times some constant. Increase the contiguity, the score will increase
ctg_L50 - Given a set of contigs, the L50 is defined as the sequence length of the shortest contig at 50% of the total genome length.
ctg_L90 - The L90 statistic is less than or equal to the L50 statistic; it is the length for which the collection of all contigs of that length or longer contains at least 90% of the sum of the lengths of all contigs.
ctg_N50 - Given a set of contigs, each with its own length, the N50 count is defined as the smallest number of contigs whose length sum makes up half of genome size.
ctg_N90 - Given a set of contigs, each with its own length, the N90 count is defined as the smallest number of contigs whose length sum makes up 90% of genome size.
ctg_logsum - Maximum contig length.
ctg_max - Maximum contig length.
ctg_powsum - Powersum of all contigs is the same as logsum except that it uses the sum of (length*(length^P)) for some power P (default P=0.25).
gap_pct - The gap size percentage of all scaffolds.
gc_avg - Average of GC content of all contigs.
gc_std - Standard deviation of GC content of all contigs.
num_aligned_reads - The sequence count number of input reads aligned to assembled contigs.
num_input_reads - The sequence count number of input reads for assembly.
scaf_L50 - Given a set of scaffolds, the L50 is defined as the sequence length of the shortest scaffold at 50% of the total genome length.
scaf_L90 - The L90 statistic is less than or equal to the L50 statistic; it is the length for which the collection of all scaffolds of that length or longer contains at least 90% of the sum of the lengths of all scaffolds.
scaf_N50 - Given a set of scaffolds, each with its own length, the N50 count is defined as the smallest number of scaffolds whose length sum makes up half of genome size.
scaf_N90 - Given a set of scaffolds, each with its own length, the N90 count is defined as the smallest number of scaffolds whose length sum makes up 90% of genome size.
scaf_bp - Total size in bp of all scaffolds.
scaf_l_gt50K - Total size in bp of all scaffolds greater than 50 KB.
scaf_logsum - The sum of the (length*log(length)) of all scaffolds, times some constant. Increase the contiguity, the score will increase
scaf_max - Maximum scaffold length.
scaf_n_gt50K - Total sequence count of scaffolds greater than 50 KB.
scaf_pct_gt50K - Total sequence size percentage of scaffolds greater than 50 KB.
scaf_powsum - Powersum of all scaffolds is the same as logsum except that it uses the sum of (length*(length^P)) for some power P (default P=0.25).
scaffolds - Total sequence count of all scaffolds.
metagenome assembly set - This property links a database object to the set of metagenome assembly activities.
metaproteomics analysis activity set - This property links a database object to the set of metaproteomics analysis activities.
metatranscriptome activity set - This property links a database object to the set of metatranscriptome analysis activities.
min_q_value - smallest Q-Value associated with the peptide sequence as provided by MSGFPlus tool
mod_date - The last date on which the database information was modified.
name - A human readable label for an entity
person value➞name - The full name of the Investgator. It should follow the format FIRST [MIDDLE NAME| MIDDLE INITIAL] LAST, where MIDDLE NAME| MIDDLE INITIAL is optional.
nmdc schema version - TODO
nom analysis activity set - This property links a database object to the set of natural organic matter (NOM) analysis activities.
object set - Applies to a property that links a database object to a set of objects. This is necessary in a json document to provide context for a list, and to allow for a single json object that combines multiple object types
objective - The scientific objectives associated with the entity. It SHOULD correspond to scientific norms for objectives field in a structured abstract.
omics processing set - This property links a database object to the set of omics processings within it.
omics type - The type of omics data
orcid - The ORICD of a person.
output_read_bases - TODO
part of - Links a resource to another resource that either logically or physically includes it.
peptide_sequence_count - count of peptide sequences grouped to the best_protein
peptide_spectral_count - sum of filter passing MS2 spectra associated with the peptide sequence within a given LC-MS/MS data file
peptide_sum_masic_abundance - combined MS1 extracted ion chromatograms derived from MS2 spectra associated with the peptide sequence from a given LC-MS/MS data file using the MASIC tool
phase - The phase for a coding sequence entity. For example, phase of a CDS as represented in a GFF3 with a value of 0, 1 or 2.
processing_institution - The organization that processed the sample.
profile image url - A url that points to an image of a person.
protein_spectral_count - sum of filter passing MS2 spectra associated with the best protein within a given LC-MS/MS data file
protein_sum_masic_abundance - combined MS1 extracted ion chromatograms derived from MS2 spectra associated with the best protein from a given LC-MS/MS data file using the MASIC tool
publications - A list of publications that are assocatiated with the entity. The publicatons SHOULD be given using an identifier, such as a DOI or Pubmed ID, if possible.
read QC analysis activity set - This property links a database object to the set of read QC analysis activities.
-
input base count - The nucleotide base count number of input reads for QC analysis.
input read count - The sequence count number of input reads for QC analysis.
output base count - After QC analysis nucleotide base count number.
output read count - After QC analysis sequence count number.
read based analysis activity set - This property links a database object to the set of read based analysis activities.
salinity_category - Categorcial description of the sample’s salinity. Examples: halophile, halotolerant, hypersaline, huryhaline
seqid - The ID of the landmark used to establish the coordinate system for the current feature.
smiles - A string encoding of a molecular graph, no chiral or isotopic information. There are usually a large number of valid SMILES which represent a given structure. For example, CCO, OCC and C(O)C all specify the structure of ethanol.
strand - The strand on which a feature is located. Has a value of ‘+’ (sense strand or forward strand) or ‘-’ (anti-sense strand or reverse strand).
study image - Links a study to one or more images.
study set - This property links a database object to the set of studies within it.
term - pointer to an ontology class
title - A name given to the entity that differs from the name/label programatically assigned to it. For example, when extracting study information for GOLD, the GOLD system has assigned a name/label. However, for display purposes, we may also wish the capture the title of the proposal that was used to fund the study.
type - An optional string that specifies the type object. This is used to allow for searches for different kinds of objects.
attribute value➞type - An optional string that specified the type of object.
functional annotation➞type - TODO
genome feature➞type - A type from the sequence ontology
-
metabolomics analysis activity➞used - The instrument used to collect the data used in the analysis
metaproteomics analysis activity➞used - The instrument used to collect the data used in the analysis
nom analysis activity➞used - The instrument used to collect the data used in the analysis
-
workflow execution activity➞was associated with - the agent/entity associated with the generation of the file
-
functional annotation➞was generated by - provenance for the annotation.
websites - A list of websites that are assocatiated with the entity.
Enums
[credit enum](credit enum.md)
[file type enum](file type enum.md)
Subsets
DataObjectSubset - Subset consisting of the data objects that either inputs or outputs of processes or workflows.
SampleSubset - Subset consisting of entities linked to the processing of samples. Currently, this subset consists of study, omics process, and biosample.
WorkflowSubset - Subset consisting of just the workflow exectution activities
Types
Built in
Bool
Decimal
ElementIdentifier
NCName
NodeIdentifier
URI
URIorCURIE
XSDDate
XSDDateTime
XSDTime
float
int
str
Defined
Boolean (Bool) - A binary (true or false) value
Bytes (int) - An integer value that corresponds to a size in bytes
Date (XSDDate) - a date (year, month and day) in an idealized calendar
Datetime (XSDDateTime) - The combination of a date and time
Decimal (Decimal) - A real number with arbitrary precision that conforms to the xsd:decimal specification
DecimalDegree (float) - A decimal degree expresses latitude or longitude as decimal fractions.
Double (float) - A real number that conforms to the xsd:double specification
ExternalIdentifier (Uriorcurie) - A CURIE representing an external identifier
Float (float) - A real number that conforms to the xsd:float specification
Integer (int) - An integer
LanguageCode (str) - A language code conforming to ISO_639-1
Ncname (NCName) - Prefix part of CURIE
Nodeidentifier (NodeIdentifier) - A URI, CURIE or BNODE that represents a node in a model.
Objectidentifier (ElementIdentifier) - A URI or CURIE that represents an object in the model.
String (str) - A character string
Time (XSDTime) - A time object represents a (local) time of day, independent of any particular day
Unit (str)
Uri (URI) - a complete URI
Uriorcurie (URIorCURIE) - a URI or a CURIE
The NMDC Metadata Standards Documentation
Introduction
This documentation provides details on the National Microbiome Data Collaborative’s (NMDC) approach to sample and data processing metadata. These are key features that drive the data search and discovery aspect of the NMDC data portal (https://microbiomedata.org/data/). If you are unfamiliar with these types of metadata (Figure 1), we recommend you begin with an Introduction to Metadata and Ontologies: Everything You Always Wanted to Know About Metadata and Ontologies (But Were Afraid to Ask) (https://doi.org/10.25979/1607365).
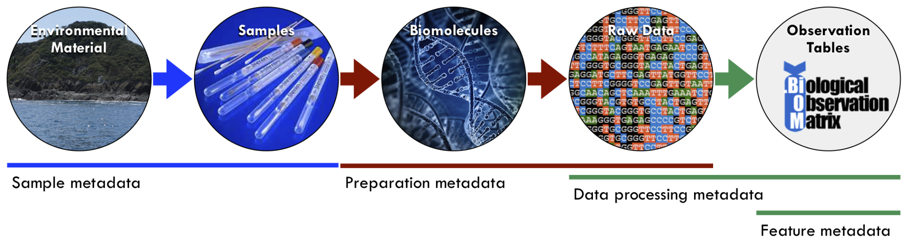
Figure 1: Microbiome metadata types: Information that contextualizes sample including its geographic location and collection date, sample preparation, data processing methods, and data products produced from a biological sample (Luke et al., 2020. Introduction to Metadata and Ontologies: Everything You Always Wanted to Know About Metadata and Ontologies (But Were Afraid to Ask). DOI: 10.25979/1607365).
All data integrated into the NMDC data portal must adhere to existing metadata standards for proper indexing and display, and to ensure accurate search results are returned. This documentation outlines the standards and ontologies that were included in the NMDC data schema, a framework that defines how data were defined and linked. For the 2019-2022 pilot initiative, the NMDC Metadata Standards Team (see the NMDC Team page) leveraged existing community-driven standards developed by the Genomics Standards Consortium (GSC), the Joint Genome Institute (JGI) Genomes Online Database (GOLD), and OBO Foundry’s Environmental Ontology (EnvO). In collaboration with these organizations, the NMDC has created a framework for mapping these standards into an interoperable framework that can be expanded to include additional standards and ontologies in the future.
Additional information on the activities by the NMDC Metadata Standards team can be found on the NMDC website at: https://microbiomedata.org/metadata/
Standards and Ontologies used by the NMDC
Sample Metadata
GSC Minimum Information about any (x) Sequence (MIxS)
The GSC has developed standards for describing genomic and metagenomic sequences, including the “minimum information about a genome sequence” (MIGS), the “minimum information about a metagenome sequence” (MIMS), and the “minimum information about a marker gene sequence” (MIMARKS). To complement this community-driven standard effort, the GSC has also developed a system for describing the environment from which a biological sample originates, as “environmental packages” and established a unified standard set of checklists through the minimum information about any (x) sequence (MIxS). MIxS provides a standardized data dictionary of sample descriptors (e.g., location, environment, elevation, altitude, depth, etc.) organized into different packages for 17 different sample environments.
To standardize how physical samples are described (i.e., sample metadata, Figure 1), the NMDC schema includes environmental descriptors from the GSC MIxS standards.
Explore how to create a MIxS-compliant sample metadata spreadsheet
Review our example spreadsheet with sample metadata that has been converted to be compliant with the MIxS Soil environment package. Note that not all non-mandatory terms from the MIxs Soil package were relevant for these example samples, and hence were omitted for clarity.
Basic sample spreadsheet (Tab 1 - before conversion to MIxS)
MIxS-compliant soil spreadsheet (Tab 2 - converted to MIxS Soil)
Explore the mandatory, unique, and shared descriptors from the MIxS Soil package
Searchable descriptors from all MIxS environmental packages - coming soon!
Learn more about all of the 17 MIxS environmental packages
Genomes Online Database (GOLD)
The JGI Genomes OnLine Database (GOLD, Mukherjee 2021) is an open-access repository of genome, metagenome, and metatranscriptome sequencing projects with their associated metadata. GOLD data are organized based on Study, Biosample/Organism, Sequencing Project and Analysis Project (Mukherjee 2017). Biosamples (defined as the physical material collected from an environment) are described using a five-level ecosystem classification path (Figure 2); the NMDC schema also uses this ecosystem classification to describe sample environments.
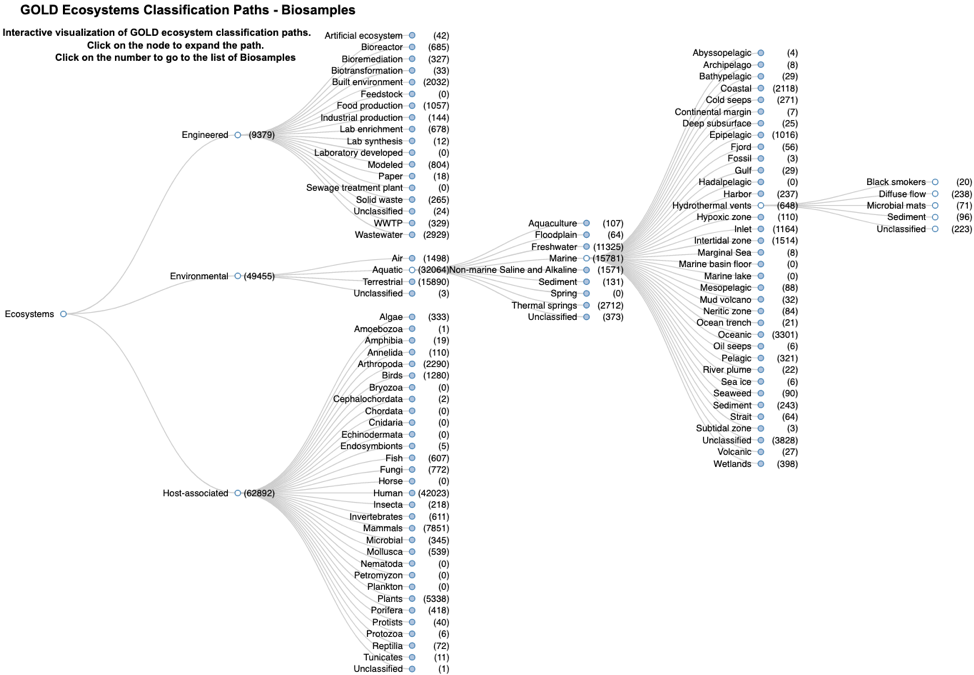
Figure 2. The GOLD five-level ecosystem classification paths (Mukherjee 2019).
Overview of the GOLD ecosystem paths
Ecosystem describes biosamples using three different broadest contexts, namely environmental, engineered, and host-associated.
Ecosystem category subdivides the ecosystem into categories, such as aquatic or terrestrial.
Ecosystem type classifies those categories into types, such as freshwater or marine, cave, desert, soil, etc.
Ecosystem subtype allows for additional environmental context or boundaries.
Specific ecosystem that describes the environment that directly influences the sample or the environmental material itself.
Explore how to map sample environments using the GOLD ecosystem classification
Learn more about the GOLD ecosystem paths using an interactive visualization tool.
Review a step-by-step example of how to assign the GOLD ecosystem classification to a lake sediment sample.
Environmental Ontology (EnvO)
The Environment Ontology (EnvO, Buttigieg 2016) is a community-led ontology that represents environmental entities such as biomes, environmental features, and environmental materials. Each EnvO term is identified using a unique resource identifier (e.g., CURIE or IRI) that resolves in a web browser. This ensures that EnvO’s terms (and their definitions) are easy to find, reference, and share amongst collaborators. It also ensures that datasets described using EnvO terms can be more easily integrated and analyzed in a reproducible manner. And since the meanings of the terms are precisely defined and accessible, humans and computers can easily connect EnvO terms across datasets.
EnvO terms are the recommended values for several of the mandatory terms in the MIxS packages, often referred to as the “MIxS triad”.
MIxS: env_broad_scale (a.k.a. Biome): The major environmental system that the sample or specimen came from. Often, the value for this term comes from EnvO’s biome hierarchy, and is similar to GOLD’s Ecosystem category.
Examples: forest biome, tropical biome, and oceanic pelagic zone biome
MIxS: env_local_scale (a.k.a. Feature): A more direct expression of the sample or specimen’s local vicinity, which likely has a significant influence on the sample or specimen. Possible values are listed in EnvO’s astronomical body part hierarchy, which is similar to GOLD’s Ecosystem type/subtype.
Examples: mountain, pond, whale fall, and karst
MIxS: env_medium (a.k.a. material): The environmental material(s) immediately surrounding your sample or specimen prior to sampling. Examples of this are found in EnvO’s environmental material hierarchy, and is similar to GOLD’s Specific ecosystem.
Examples: sediment, soil, water, and air
Explore how to map sample environments using the EnvO ecosystem classification
Review a step-by-step example of how to assign EnvO terms to an oligotrophic lake sediment sample below.
|
env_broad_scale (Biome) Using EnvO biome categories, aquatic is appropriate. However, since the EnvO is a hierarchical system, the aquatic biome has two sub-categories: freshwater and marine biomes. The freshwater biome is further divided into freshwater lake biome and freshwater river biome. Therefore, for a lake sediment sample, freshwater lake biome is the appropriate EnvO biome category. |

|
|
env_local_scale (Feature) Next, we describe the local environmental feature in the vicinity of and likely having a strong causal influence on the sample. Using the EnvO astronomical body part categories, we step through the relevant categories (see figure on the right) until we reach the EnvO term oligotrophic lake. |

|
|
env_medium (Material) Finally, since the sample is oligotrophic lake sediment, the EnvO environmental material could be assigned sediment. But because the EnvO hierarchy provides sub-categories within sediment, the environmenta material will be assigned lake sediment. |

|
Therefore, the EnvO triad for oligotrophic lake sediment is:
Env_broad_scale: freshwater lake biome [ENVO_01000252]
Env_local_scale: oligotrophic lake [ENVO_01000774]
Env_medium: lake sediment [ENVO_00000546]
Classifying samples with GOLD and MIxS/EnvO
The five-level GOLD ecosystem classification path and EnvO triad each have unique advantages in describing the environmental context of a biosample. The NMDC leverages the strengths of both the GOLD ecosystem classification path and MIxS/EnvO triad. The assignment of MIxS/EnvO triad for the biosamples currently in the NMDC data portal was achieved through a manual curation process using various metadata fields of GOLD biosamples fields, such as name, description, habitat, sample collection site, identifier, ecosystem classification path, and study description. The NMDC team is currently working on exploring solutions for automated mapping between GOLD and MIxS/EnvO.
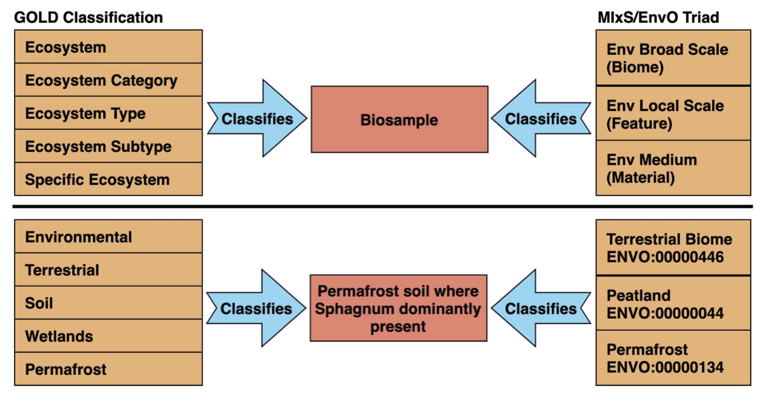
Figure 3: Mapping between the MIxS/EnvO triad and the GOLD ecosystem classification enables integration of sample environments defined with GOLD and MIxS/EnvO.
Data Processing Metadata
In addition, the NMDC is adopting the MIxS standards for sequence data types (e.g., sequencing method, pcr primers and conditions, etc.), and are building on previous efforts by the Proteomics Standards Initiative and Metabolomics Standards Initiative to develop standards and controlled vocabularies for mass spectrometry data types (e.g., ionization mode, mass resolution, scan rate, etc.). Additional details on the processing metadata are coming soon.
Overview of the NMDC Data Schema
The NMDC has developed a normalized metadata schema (available in the NMDC GitHub) for representing studies, samples, relationships between samples, and associated data objects. The schema is organized into object classes, which act as nodes. Each class has associated slots, which are fields that contain metadata that describe the object. For more in-depth information, full documentation of the NMDC schema can be found here.
For the NMDC pilot, a python toolkit for generating NMDC-compliant JavaScript Object Notation (JSON) objects was developed to create ETL (Extract-Transform-Load) software to ingest metadata from the DOE User Facilities. Read more about the data in the NMDC pilot here.
Validating json objects against the NMDC schema
This document assumes knowledge of JSON. It also assumes rudimentary familiarity with JSON-Schema but don’t worry if you are not an expert on this.
We can conceive of validation of a piece of JSON at two levels
The JSON should be syntactically correct JSON
The JSON should conform to the NMDC schema
Syntactically correct JSON
It is crucial that the JSON is syntactically valid, otherwise it can’t even be schema-validated.
There are a variety of ways to check for this. We recommend using jsonschema to validate this, see below.
NOTE: all NMDC JSON-producing tools, libraries, or scripts SHOULD use a standard json library. If you are using a robust standard json library, your output is practically guaranteed to be syntactically valid JSON.
It is strongly recommended that you do NOT generate JSON by methods such as directly manipulating json strings or printing directly. This is guaranteed to be fragile/non-robust. Even if your code works now, it is certain it will fail later and produce incorrect JSON.
For Python, there is only one choice:
https://docs.python.org/3/library/json.html
If you are not using this, you should
Schema validation
The JSON-Schema for NMDC is maintained in this github repo, under jsonschema/nmdc.schema.json
Note that the JSON-Schema is generated from a higher level YAML representation, using a modeling framework called linkML. See the README for details. For understanding the schema, you may be better looking at the auto-generated docs. However, for computational conformance, the JSON-Schema is what is should be used.
There are a variety of json schema validators, these will give the same results. There are web playgrounds for this. But for simplicity we recommend the Python jsonschema package
To install:
pip install jsonschema
Assume you have a file MYFILE that is json intended to conform
jsonschema -i /PATH/TO/MYFILE.json jsonschema/nmdc.schema.json
If the json is valid, there will be no output and the script will pass. If there are problems these will be reported.
You can try this with some ready-made examples in this repo:
jsonschema -i examples/nmdc-01.json jsonschema/nmdc.schema.json
Note: nmdc.schema.json describes each model object, its required attributes and attribute types. The examples themselves use JSON notation to allow multiple instances of the objects in the JSON schema, to be submitted in one file.
You can also use the jsonschema library to validate directly from within your python.
What to do if your JSON does not validate
There are 3 possibilities:
Your json is good, and the schema needs to be extended or modified to account
you need to modify the json to conform
some other odd bug somewhere
For 1, you can go right ahead and make PR on the schema yaml. However, if you are not comfortable doing this then you can get help from one of the schema developers. We recommend filing a new ticket explaining the issue.
For 2, this is upon you to fix this, however debugging can be aided in pulling out single instances of your model objects, and verifying that you are creating valid JSON (ie: paste one instance of your object into https://jsonlint.com/ or tools like it to verify its syntax). Another common issue is that you might have incorrect syntax for grouping many instances of a JSON object into an array. Using a small subsample of your data and an online linter as above, can aide in debugging this. Sometimes the validation can complain about invalid syntax if the attribute of an instance object disagrees with the schema’s typing (ie: you have an integer where a string is expected).
NMDC Producer SOP
It is expected that different providers of JSON within the NMDC take responsibility for validating their JSON. Aim1 can help with any problems.
Currently not all providers of information to NMDC provide JSON - for example, GOLD is provided as database dumps, and an ETL process transforms this into JSON. In future we would like to move towards a situation where all information is provided as JSON.
Identifiers in NMDC
Identifiers are crucial for the NMDC, both for any data objects created (aka minted) and for any external objects referenced
Examples of entities that require identifiers:
Samples
Data objects (e.g. sequence files)
Taxa (e.g. NCBITaxon or GTDB)
Genes, Proteins
Sequences (e.g. genome/transcriptome)
Ontology terms and other descriptors
functional orthologs, e.g. KEGG.orthology (KO) terms
pathways, e.g. KEGG.pathway, MetaCyc, GO
reactions/activities: KEGG, MetaCyc
chemical entities: CHEBI, CHEMBL, INCHI, …
sequence feature types: SO, Rfam
Identifiers should be:
Permanent
Unique
Resolvable
Opaque
See McMurry et al, PMID:28662064 for more desiderata.
CURIEs - prefixed IDs
Following McMurry et al we adopt the use of prefixed identifiers
The syntax is:
Prefix:LocalId
Examples:
GO:0008152
BIOSAMPLE:SAMEA2397676
DOI:10.1038/nbt1156
These prefixed identifiers are also known as CURIEs (Compact URIs). There is a W3C specification for these
All prefixes should be registered with a standard identifier prefix system. These include:
http://n2t.net
http://identifiers.org
http://obofoundry.org
Examples
INSDC BioSamples
Registry entry: https://registry.identifiers.org/registry/biosample
Example ID/CURIE: BIOSAMPLE:SAMEA2397676
Resolving via identifiers.org: https://identifiers.org/BIOSAMPLE:SAMEA2397676
Resolving via nt2.net: http://n2t.net/BIOSAMPLE:SAMEA2397676
GOLD identifiers
https://registry.identifiers.org/registry/gold
Example ID: GOLD:Gp0119849
Resolving via identifiers.org: https://identifiers.org/GOLD:Gp0119849
identifiers for ontology terms and function descriptors
Most of the ontologies we use are in OBO. All OBO IDs are prefixed using the ontology ID space. The list of ID spaces can be found on http://obofoundry.org
For example the ID/CURIE ENVO:00002007 represents the class sediment and is expanded to a URI of http://purl.obolibrary.org/obo/ENVO_00002007
KEGG
KEGG is actually a set of databases, each with its own prefix, usually of form KEGG.$database, e.g.
KEGG.ORTHOLOGY (aka KO), e.g. KEGG.ORTHOLOGY:K00001
KEGG.COMPOUND, e.g. KEGG.COMPOUND:C12345
Recommended IDs for use within NMDC
The NMDC schema is annotated with the set of IDs that are allowed to act as primary keys for instances of each class.
For example the class OrthologyGroup has a description of the IDs allowed on the class web page, the first listed is KEGG.ORTHOLOGY
The underlying yaml looks like this:
orthology group:
is_a: functional annotation term
description: >-
A set of genes or gene products in which all members are orthologous
id_prefixes:
- KEGG.ORTHOLOGY ## KO number
- EGGNOG
- PFAM
- TIGRFAM
- SUPFAM
- PANTHER.FAMILY
exact_mappings:
- biolink:GeneFamily
The full URLs for each is in the jsonld context file
IDs minted for use within NMDC
Note that NMDC schema mandates IDs for most objects. These always have the field name id
Reuse vs minting new IDs
We try to reuse IDs as far as possible. For example, for any sample already in GOLD, we use the GOLD sample identifier, e.g. GOLD:Gb…..
IDs generated during workflows
This section is in progress. See https://github.com/microbiomedata/nmdc-metadata/issues/195
All instances of OmicsProcessing have IDs. The policy for ID depends on the provider.
Currently metagenomics omics objects look like this:
id: "gold:Gp0108335"
name: "Thawing permafrost microbial communities from the Arctic, studying carbon transformations - Permafrost 712P3D"
has_input:
- "gold:Gb0108335"
part_of:
- "gold:Gs0112340"
has_output:
- "jgi:551a20d30d878525404e90d5"
omics_type: Metagenome
type: "nmdc:OmicsProcessing"
add_date: "30-OCT-14 12.00.00.000000000 AM"
mod_date: "22-MAY-20 06.13.12.927000000 PM"
ncbi_project_name: "Thawing permafrost microbial communities from the Arctic, studying carbon transformations - Permafrost 712P3D"
processing_institution: "Joint Genome Institute"
principal_investigator_name: "Virginia Rich"
note that we use re-using the GOLD ID rather than minting a new one
the linked data object uses a jgi prefix and an md5 hash
id: "jgi:551a20d30d878525404e90d5"
name: "8871.1.114459.GCCAAT.fastq.gz"
description: "Raw sequencer read data"
file_size_bytes: 17586370657
type: "nmdc:DataObject"
note that currently jgi is not registered and thus the ID is not resolvable
Currently metaproteomics omics objects look like this:
id: "emsl:404590"
name: "FECB_21_5093B_01_23Dec14_Tiger_14-11-12"
description: "High res MS with low res CID MSn"
part_of:
- "gold:Gs0110132"
has_output:
- "emsl:output_404590"
omics_type: Proteomics
type: "nmdc:OmicsProcessing"
instrument_name: "VOrbiETD03"
processing_institution: "Environmental Molecular Sciences Lab"
this is suboptimal; emsl is not yet registered, and it’s not clear that the integer is unique within emsl, let alone the nmdc subset
the output data objects are formed from these:
id: "emsl:output_404590"
name: "output: FECB_21_5093B_01_23Dec14_Tiger_14-11-12"
description: "High res MS with low res CID MSn"
file_size_bytes: 503296678
type: "nmdc:DataObject"
the data objects use hashes (md5) prefixed with nmdc:
name: "404590_resultant.tsv"
description: "Aggregation of analysis tools{MSGFplus, MASIC} results"
file_size_bytes: 10948480
type: "nmdc:DataObject"
id: "nmdc:e0c70280a7a23c7c5cc1e589f72e896e"
note nmdc is not yet registered
Both metaG and metaT analyses produce GFF3 files. See issue 184 for more on how the GFF is modeled.
The main entity we care about in these is the [gene product] https://microbiomedata.github.io/nmdc-metadata/docs/GeneProduct) ID (usually a protein), this is what functional annotation hangs off.
This is typically a protein encoded by a CDS, e.g.
Ga0185794_41 GeneMark.hmm-2 v1.05 CDS 48 1037 56.13 + 0 ID=Ga0185794_41_48_1037;translation_table=11;start_type=ATG;product=5-methylthioadenosine/S-adenosylhomocysteine deaminase;product_source=KO:K12960;cath_funfam=3.20.20.140;cog=COG0402;ko=KO:K12960;ec_number=EC:3.5.4.28,EC:3.5.4.31;pfam=PF01979;superfamily=51338,51556
Currently we are prefixing the ID field in GFF with nmdc, e.g. nmdc:Ga0185794_41_48_1037 as the protein ID
When converting col9 we ensure that each ID is correctly prefixed. So for example, we use KEGG.OTHOLOGY:K12960 not KO:K12960 as the former is the official prefix according to KEGG and identifiers.org
We will also later need a policy for IDs for the sequences in col1 (ie genome or transcript), please return later for more details…
MIxS term identifiers
We are working with the GSC to provide permanent IDs for MIxS terms. Note these terms are schema-level rather than data-level.
Please check this section later
For now we place these in the nmdc namespaces, e.g
nmdc:alt
Identifier mapping
Please check this section later
Identifiers and semantic web URIs
We produce a JSON-LD context with the schema:
When this is combined with schema-conformant JSON, RDF can be automatically created using the intended URIs
MIxS Soil Package
The MIxS Soil Package contains a list of 145 descriptors to describe the soil sample taken from various environments including soil from, cropland, dryland, forest, grassland soil, coastal sand dune, permafrost soil. These 145 descriptors have been provided in different sections namely soil, nucleic acid sequence source, environment, sequencing, investigation and MIxS extension. We have grouped these descriptors into mandatory descriptors, unique descriptors and other descriptors (non mandatory and non unique).
Some examples of biosamples described using MIxS-Soil package (v5) terms:
https://www.ncbi.nlm.nih.gov/biosample/SAMN07125075
https://www.ncbi.nlm.nih.gov/biosample/SAMN08902834
Mandatory descriptors of MIxS Soil packages are:
The MIxS soil package has 12 mandatory descriptors including ‘depth’ and ‘elevation’. These 12 mandatory descriptors with descriptor name, definition, section of the MIxS package, expected value, value syntax for all of the descriptors and preferred unit and example value when available are listed below.
investigation_type - Nucleic Acid Sequence Report is the root element of all MIGS/MIMS compliant reports as standardized by Genomic Standards Consortium. This field is either eukaryote,bacteria,virus,plasmid,organelle, metagenome,mimarks-survey, mimarks-specimen, metatranscriptome, single amplified genome, metagenome-assembled genome, or uncultivated viral genome.
Section : investigation
Expected value : eukaryote, bacteria_archaea, plasmid, virus, organelle, metagenome,mimarks-survey, mimarks-specimen, metatranscriptome, single amplified genome, metagenome-assembled genome, or uncultivated viral genomes
Value syntax : [eukaryote|bacteria_archaea|plasmid|virus|organelle|metagenome|metatranscriptome|mimarks-survey|mimarks-specimen|misag|mimag|miuvig]
Example : metagenomeproject_name - Name of the project within which the sequencing was organized.
Section : investigation
Expected value :
Value syntax : {text}The project name in the NMDC follows standardized metagenome naming scheme as per the Genomes Online Database (GOLD) that can be accessed fromhttps://gold.jgi.doe.gov/resources/Standardized_Metagenome_Naming.pdf
The following four metadata are used in the naming of the project:
[Habitat] [Type of communities] [ Location, including the country/ocean] – [Identifier]
For example, for the following metadata:
Habitat: Permafrost
COMMUNITY: microbial communities
GEOGRAPHIC_LOCATION: Sweden: Stordalen mire
Sample_Identifier: 20120800_S1X
Project name for metagenome would be:
Permafrost microbial communities from Stordalen mire, Sweden - 20120800_S1X.
Project name for Metatranscriptome would be:
Metatranscriptome of permafrost microbial communities from Stordalen mire, Sweden - 20120800_S1X
lat_lon - The geographical origin of the sample as defined by latitude and longitude. The values should be reported in decimal degrees and in WGS84 system.
Section : environment
Expected value : decimal degrees
Value syntax : {float} {float}
Example : 50.586825 6.408977geo_loc_name - The geographical origin of the sample as defined by the country or sea name followed by specific region name. Country or sea names should be chosen from the INSDC country list (http://insdc.org/country.html), or the GAZ ontology that can be accessed from http://www.ontobee.org/ontology/GAZ or http://purl.bioontology.org/ontology/GAZ.
Section : environment
Expected value : country or sea name (INSDC or GAZ);region(GAZ);specific location name
Value syntax : {term};{term};{text}
Example : Germany;North Rhine-Westphalia;Eifel National Parkcollection_date - The time of sampling, either as an instance (single point in time) or interval. In case no exact time is available, the date/time can be right truncated i.e. all of these are valid times: 2008-01-23T19:23:10+00:00; 2008-01-23T19:23:10; 2008-01-23; 2008-01; 2008; Except: 2008-01; 2008 all are ISO8601 compliant.
Section : environment
Expected value : date and time
Value syntax : {timestamp}
Example : 2018-05-11T10:00:00+01:00env_broad_scale - The broad-scale environmental context of MIxS uses terminologies from Environment Ontology (EnvO). EnvO describes the broad-scale environmental context as environmental systems / biomes to which resident ecological communities have evolved adaptations. Biome possesses a degree of spatial and temporal stability that has allowed at least some of its constituent communities to adapt. In this field, report which major environmental system your sample or specimen came from. The systems identified should have a coarse spatial grain, to provide the general environmental context of where the sampling was
done (e.g. were you in the desert or a rainforest?).
Some of the broad-scale environmental context terms from EnvO that can be used for soil biosamples are, terrestrial biome, anthropogenic terrestrial biome, desert biome, cropland biome, forest biome, mixed forest biome, grassland biome, tropical biome, tropical grassland biome, tundra biome and urban biome.
We recommend using subclasses of ENVO’s biome class: Biome class represents http://purl.obolibrary.org/obo/ENVO_00000428.
Section : environment
Expected value : Add terms that identify the major environment type(s) where your sample was collected. Recommend subclasses of biome [ENVO:00000428]. Format for single term: termLabel [termID],Format for multiple terms: termLabel [termID]|termLabel [termID]|termLabel [termID].Value syntax : {termLabel} {[termID]}
Example:annotating soil from permafrost: terrestrial biome [ENVO_00000446] or
soil from meadow: grassland biome [ENVO_01000177]
terrestrial biome [ENVO_00000446]|urban biome[ENVO_01000249]
env_local_scale - The local environmental context of MIxS uses terminologies from Environment Ontology (EnvO). EnvO describes the local environmental context as environmental features that are in the vicinity of and have a strong causal influence on the entity; in this field, report the entity or entities which are in your sample or specimen’s local vicinity and which you believe have significant causal influences on your sample or specimen. Some of the MIxS local environmental context terms from EnvO that can be used describe soil feature are: agricultural field, desert, flood plain, garden, hill, paddy field and river bank etc. The MIxS local environmental context terms given in ENVO that are of smaller spatial grain than your entry for env_broad_scale.
If needed, request new terms on the ENVO tracker, identified here: http://www.obofoundry.org/ontology/envo.html.
Section : environment
Expected value : Add terms that identify environmental entities having causal influences upon the entity at time of sampling. Format for single term: termLabel [termID]; Format for multiple terms: termLabel [termID]|termLabel [termID]|termLabel [termID].Value syntax : {termLabel} {[termID]}
Example:
annotating local environmental context of soil from permafrost active layer: active permafrost layer [ENVO_04000009] or
soil from a biosphere reserve: biosphere reserve [ENVO_00000376]
agricultural field[ENVO_00000114]|banana plantation[ENVO_00000161]
env_medium - The MIxS environmental medium context terms uses terminologies from Environment Ontology (EnvO). EnvO describes the environmental medium/material context terms as those terms that refers to masses, volumes, or other portions of some medium included in an environmental system; environmental material that is the substance surrounding or partially surrounding the entity.
Some of the MIxS env_medium terms from EnvO that can be used describe soil biosamples are: agricultural soil, bulk soil, burned soil, eucalyptus forest soil, forest soil, farm soil, fertilized soil, forest soil, garden soil, grassland soil, greenhouse soil, heat stressed soil, meadow soil, peat soil, soil, spruce forest soil, surface soil etc.
In this field, report which environmental material or materials (pipe separated) immediately surrounded your sample or specimen prior to sampling, using one or more subclasses of ENVO’s environmental material class: http://purl.obolibrary.org/obo/ENVO_00010483.
Section : environment
Expected value : Add terms that identify the material displaced by the entity at time of sampling. Recommend subclasses of environmental material [ENVO:00010483]. Multiple terms can be separated by pipes e.g.: estuarine waterFormat (one term): termLabel [termID];
Format (multiple terms): termLabel [termID]|termLabel [termID]|termLabel [termID].
Value syntax : {termLabel} {[termID]}
Example:Annotating env_medium (environmental medium context terms) of meadow soil: meadow soil [ENVO_00005761].
When there are multiple terms, agricultural soil [ENVO_00002259]|bulk soil [ENVO_00005802]|oil contaminated soil [ENVO_00002875]
depth - Depth is defined as the vertical distance below local surface, e.g. For sediment or soil samples depth is measured from sediment or soil surface, respectively. Depth can be reported as an interval for subsurface samples.
Section : soil
Expected value : measurement value
Preferred unit : meter
Value syntax : {float} {unit}
Example : 10 meterelev - Elevation of the sampling site is its height above a fixed reference point, most commonly the mean sea level. Elevation is mainly used when referring to points on the earth’s surface, while altitude is used for points above the surface, such as an aircraft in flight or a spacecraft in orbit.
Section : soil
Expected value : measurement value
Preferred unit : meter
Value syntax : {float} {unit}
Example : 100 metersubmitted_to_insdc - Depending on the study (large-scale e.g. done with next generation sequencing technology, or small-scale) sequences have to be submitted to SRA (Sequence Read Archive), DRA (DDBJ Read Archive) or via the classical Webin/Sequin systems to Genbank, ENA and DDBJ. Although this field is mandatory, it is meant as a self-test field, therefore it is not necessary to include this field in contextual data submitted to databases.
Section : investigation
Expected value : boolean
Value syntax : {boolean}
Example : yesseq_meth - Sequencing method used; e.g. Sanger, pyrosequencing, ABI-solid.
Section : sequencing
Expected value : enumeration
Value syntax : [MinION|GridION|PromethION|454 GS|454 GS 20|454 GS FLX|454 GS FLX+|454 GS FLX Titanium|454 GS Junior|Illumina Genome Analyzer|Illumina Genome Analyzer II|Illumina Genome Analyzer IIx|Illumina HiSeq 4000|Illumina HiSeq 3000|Illumina HiSeq 2500|Illumina HiSeq 2000|Illumina HiSeq 1500|Illumina HiSeq 1000|Illumina HiScanSQ|Illumina MiSeq|Illumina HiSeq X Five|Illumina HiSeq X Ten|Illumina NextSeq 500|Illumina NextSeq 550|AB SOLiD System|AB SOLiD System 2.0|AB SOLiD System 3.0|AB SOLiD 3 Plus System|AB SOLiD 4 System|AB SOLiD 4hq System|AB SOLiD PI System|AB 5500 Genetic Analyzer|AB 5500xl Genetic Analyzer|AB 5500xl-W Genetic Analysis System|Ion Torrent PGM|Ion Torrent Proton|Ion Torrent S5|Ion Torrent S5 XL|PacBio RS|PacBio RS II|Sequel|AB 3730xL Genetic Analyzer|AB 3730 Genetic Analyzer|AB 3500xL Genetic Analyzer|AB 3500 Genetic Analyzer|AB 3130xL Genetic Analyzer|AB 3130 Genetic Analyzer|AB 310 Genetic Analyzer|BGISEQ-500]
Example : Illumina HiSeq 1500
Unique descriptors (46) in MIxS Soil package
The MIxS Soil package has 46 unique descriptors when compared with other MIxS packages. Name, definition, section of the MIxS package, expected value, value syntax for all of these descriptors and preferred unit and example value when available are listed below.
agrochem_addition - Addition of fertilizers, pesticides, etc. - amount and time of applications.
Section : soil
Expected value : agrochemical name;agrochemical amount;timestamp
Preferred unit : gram, mole per liter, milligram per liter
Value syntax : {text};{float} {unit};{timestamp}
Example : roundup;5 milligram per liter;2018-06-21al_sat - Aluminum saturation (esp. For tropical soils).
Section : soil
Expected value : measurement value
Preferred unit : percentage
Value syntax : {float} {unit}al_sat_meth - Reference or method used in determining Al saturation.
Section : soil
Expected value : PMID,DOI or URL
Value syntax : {PMID}|{DOI}|{URL}annual_precpt - The average of all annual precipitation values known, or an estimated equivalent value derived by such methods as regional indexes or Isohyetal maps. .
Section : soil
Expected value : measurement value
Preferred unit : millimeter
Value syntax : {float} {unit}annual_temp - Mean annual temperature.
Section : soil
Expected value : measurement value
Preferred unit : degree Celsius
Value syntax : {float} {unit}
Example : 12.5 degree Celsiuscrop_rotation - Whether or not crop is rotated, and if yes, rotation schedule.
Section : soil
Expected value : crop rotation status;schedule
Value syntax : {boolean};{Rn/start_time/end_time/duration}
Example : yes;R2/2017-01-01/2018-12-31/P6Mcur_land_use - Present state of sample site.
Section : soil
Expected value : enumeration
Value syntax : [cities|farmstead|industrial areas|roads/railroads|rock|sand|gravel|mudflats|salt flats|badlands|permanent snow or ice|saline seeps|mines/quarries|oil waste areas|small grains|row crops|vegetable crops|horticultural plants (e.g. tulips)|marshlands (grass,sedges,rushes)|tundra (mosses,lichens)|rangeland|pastureland (grasslands used for livestock grazing)|hayland|meadows (grasses,alfalfa,fescue,bromegrass,timothy)|shrub land (e.g. mesquite,sage-brush,creosote bush,shrub oak,eucalyptus)|successional shrub land (tree saplings,hazels,sumacs,chokecherry,shrub dogwoods,blackberries)|shrub crops (blueberries,nursery ornamentals,filberts)|vine crops (grapes)|conifers (e.g. pine,spruce,fir,cypress)|hardwoods (e.g. oak,hickory,elm,aspen)|intermixed hardwood and conifers|tropical (e.g. mangrove,palms)|rainforest (evergreen forest receiving >406 cm annual rainfall)|swamp (permanent or semi-permanent water body dominated by woody plants)|crop trees (nuts,fruit,christmas trees,nursery trees)]
Example : coniferscur_vegetation - Vegetation classification from one or more standard classification systems, or agricultural crop.
Section : soil
Expected value : current vegetation type
Value syntax : {text}cur_vegetation_meth - Reference or method used in vegetation classification .
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}drainage_class - Drainage classification from a standard system such as the USDA system.
Section : soil
Expected value : enumeration
Value syntax : [very poorly|poorly|somewhat poorly|moderately well|well|excessively drained]
Example : wellextreme_event - Unusual physical events that may have affected microbial populations.
Section : soil
Expected value : date
Value syntax : {timestamp}extreme_salinity - Measured salinity .
Section : soil
Expected value : measurement value
Preferred unit : millisiemens per meter
Value syntax : {float} {unit}fao_class - Soil classification from the FAO World Reference Database for Soil Resources. The list can be found at http://www.fao.org/nr/land/sols/soil/wrb-soil-maps/reference-groups.
Section : soil
Expected value : enumeration
Value syntax : [Acrisols|Andosols|Arenosols|Cambisols|Chernozems|Ferralsols|Fluvisols|Gleysols|Greyzems|Gypsisols|Histosols|Kastanozems|Lithosols|Luvisols|Nitosols|Phaeozems|Planosols|Podzols|Podzoluvisols|Rankers|Regosols|Rendzinas|Solonchaks|Solonetz|Vertisols|Yermosols]
Example : Luvisolsfire - Historical and/or physical evidence of fire.
Section : soil
Expected value : date
Value syntax : {timestamp}flooding - Historical and/or physical evidence of flooding.
Section : soil
Expected value : date
Value syntax : {timestamp}heavy_metals - Heavy metals present and concentrations any drug used by subject and the frequency of usage; can include multiple heavy metals and concentrations.
Section : soil
Expected value : heavy metal name;measurement value
Preferred unit : microgram per gram
Value syntax : {text};{float} {unit}heavy_metals_meth - Reference or method used in determining heavy metals.
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}horizon - Specific layer in the land area which measures parallel to the soil surface and possesses physical characteristics which differ from the layers above and beneath.
Section : soil
Expected value : enumeration
Value syntax : [O horizon|A horizon|E horizon|B horizon|C horizon|R layer|Permafrost]
Example : A horizonhorizon_meth - Reference or method used in determining the horizon.
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}link_addit_analys - Link to additional analysis results performed on the sample.
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}link_class_info - Link to digitized soil maps or other soil classification information.
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}link_climate_info - Link to climate resource.
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}local_class - Soil classification based on local soil classification system.
Section : soil
Expected value : local classification name
Value syntax : {text}local_class_meth - Reference or method used in determining the local soil classification .
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}microbial_biomass - The part of the organic matter in the soil that constitutes living microorganisms smaller than 5-10 micrometer. If you keep this, you would need to have correction factors used for conversion to the final units.
Section : soil
Expected value : measurement value
Preferred unit : ton, kilogram, gram per kilogram soil
Value syntax : {float} {unit}microbial_biomass_meth - Reference or method used in determining microbial biomass.
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}ph_meth - Reference or method used in determining ph.
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}pool_dna_extracts - Indicate whether multiple DNA extractions were mixed. If the answer yes, the number of extracts that were pooled should be given.
Section : soil
Expected value : pooling status;number of pooled extracts
Value syntax : {boolean};{integer}
Example : yes;5previous_land_use - Previous land use and dates.
Section : soil
Expected value : land use name;date
Value syntax : {text};{timestamp}previous_land_use_meth - Reference or method used in determining previous land use and dates.
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}profile_position - Cross-sectional position in the hillslope where sample was collected.sample area position in relation to surrounding areas.
Section : soil
Expected value : enumeration
Value syntax : [summit|shoulder|backslope|footslope|toeslope]
Example : summitsalinity_meth - Reference or method used in determining salinity.
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}season_precpt - The average of all seasonal precipitation values known, or an estimated equivalent value derived by such methods as regional indexes or Isohyetal maps. .
Section : soil
Expected value : measurement value
Preferred unit : millimeter
Value syntax : {float} {unit}season_temp - Mean seasonal temperature.
Section : soil
Expected value : measurement value
Preferred unit : degree Celsius
Value syntax : {float} {unit}
Example : 18 degree Celsiussieving - Collection design of pooled samples and/or sieve size and amount of sample sieved.
Section : soil
Expected value : design name and/or size;amount
Value syntax : {{text}|{float} {unit}};{float} {unit}slope_aspect - The direction a slope faces. While looking down a slope use a compass to record the direction you are facing (direction or degrees); e.g., nw or 315 degrees. This measure provides an indication of sun and wind exposure that will influence soil temperature and evapotranspiration.
Section : soil
Expected value : measurement value
Preferred unit : degree
Value syntax : {float} {unit}slope_gradient - Commonly called ‘slope’. The angle between ground surface and a horizontal line (in percent). This is the direction that overland water would flow. This measure is usually taken with a hand level meter or clinometer.
Section : soil
Expected value : measurement value
Preferred unit : percentage
Value syntax : {float} {unit}soil_type - Soil series name or other lower-level classification.
Section : soil
Expected value : soil type name
Value syntax : {text}soil_type_meth - Reference or method used in determining soil series name or other lower-level classification.
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}store_cond - Explain how and for how long the soil sample was stored before DNA extraction.
Section : soil
Expected value : storage condition type;duration
Value syntax : {text};{duration}
Example : -20 degree Celsius freezer;P2Y10Dtexture - The relative proportion of different grain sizes of mineral particles in a soil, as described using a standard system; express as % sand (50 um to 2 mm), silt (2 um to 50 um), and clay (<2 um) with textural name (e.g., silty clay loam) optional..
Section : soil
Expected value : measurement value
Value syntax : {float} {unit}texture_meth - Reference or method used in determining soil texture.
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}tillage - Note method(s) used for tilling.
Section : soil
Expected value : enumeration
Value syntax : [drill|cutting disc|ridge till|strip tillage|zonal tillage|chisel|tined|mouldboard|disc plough]
Example : chiseltot_nitro_content_meth - Reference or method used in determining the total nitrogen.
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}tot_org_c_meth - Reference or method used in determining total organic carbon.
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}water_content_soil_meth - Reference or method used in determining the water content of soil.
Section : soil
Expected value : PMID,DOI or url
Value syntax : {PMID}|{DOI}|{URL}
Other descriptors (non mandatory and non-unique descriptors) from MIxS Soil package
The MIxS Soil package has 89 descriptors that can also be found/used in other MIxS environmental packages. Name, definition, section of the MIxS package, expected value, value syntax for all of these descriptors and preferred unit and example value when available are listed below.
16s_recover - Can a 16S gene be recovered from the submitted SAG or MAG?.
Section : sequencing
Expected value : boolean
Value syntax : {boolean}
Example : yes
16s_recover_software - Tools used for 16S rRNA gene extraction.
Section : sequencing
Expected value : names and versions of software(s), parameters used
Value syntax : {software};{version};{parameters}
Example : rambl;v2;default parameters
adapters - Adapters provide priming sequences for both amplification and sequencing of the sample-library fragments. Both adapters should be reported; in uppercase letters.
Section : sequencing
Expected value : adapter A and B sequence
Value syntax : {dna};{dna}
Example : AATGATACGGCGACCACCGAGATCTACACGCT;CAAGCAGAAGACGGCATACGAGAT
annot - Tool used for annotation, or for cases where annotation was provided by a community jamboree or model organism database rather than by a specific submitter.
Section : sequencing
Expected value : name of tool or pipeline used, or annotation source description
Value syntax : {text}
Example : prokka
assembly_name - Name/version of the assembly provided by the submitter that is used in the genome browsers and in the community.
Section : sequencing
Expected value : name and version of assembly
Value syntax : {text} {text}
Example : HuRef, JCVI_ISG_i3_1.0
assembly_qual - The assembly quality category is based on sets of criteria outlined for each assembly quality category. For MISAG/MIMAG; Finished: Single, validated, contiguous sequence per replicon without gaps or ambiguities with a consensus error rate equivalent to Q50 or better. High Quality Draft:Multiple fragments where gaps span repetitive regions. Presence of the 23S, 16S and 5S rRNA genes and at least 18 tRNAs. Medium Quality Draft:Many fragments with little to no review of assembly other than reporting of standard assembly statistics. Low Quality Draft:Many fragments with little to no review of assembly other than reporting of standard assembly statistics. Assembly statistics include, but are not limited to total assembly size, number of contigs, contig N50/L50, and maximum contig length. For MIUVIG; Finished: Single, validated, contiguous sequence per replicon without gaps or ambiguities, with extensive manual review and editing to annotate putative gene functions and transcriptional units. High-quality draft genome: One or multiple fragments, totaling ≥ 90% of the expected genome or replicon sequence or predicted complete. Genome fragment(s): One or multiple fragments, totalling < 90% of the expected genome or replicon sequence, or for which no genome size could be estimated.
Section : sequencing
Expected value : enumeration
Value syntax : [Finished genome|High-quality draft genome|Medium-quality draft genome|Low-quality draft genome|Genome fragment(s)]
Example : High-quality draft genome
assembly_software - Tool(s) used for assembly, including version number and parameters.
Section : sequencing
Expected value : name and version of software, parameters used
Value syntax : {software};{version};{parameters}
Example : metaSPAdes;3.11.0;kmer set 21,33,55,77,99,121, default parameters otherwise
bin_param - The parameters that have been applied during the extraction of genomes from metagenomic datasets.
Section : sequencing
Expected value : enumeration
Value syntax : [homology search|kmer|coverage|codon usage|combination]
Example : coverage and kmer
bin_software - Tool(s) used for the extraction of genomes from metagenomic datasets.
Section : sequencing
Expected value : enumeration
Value syntax : [metabat|maxbin|concoct|groupm|esom|metawatt|combination|other]
Example : concoct and maxbin
biotic_relationship - Description of relationship(s) between the subject organism and other organism(s) it is associated with. E.g., parasite on species X; mutualist with species Y. The target organism is the subject of the relationship, and the other organism(s) is the object.
Section : nucleic acid sequence source
Expected value : enumeration
Value syntax : [free living|parasitism|commensalism|symbiotic|mutualism]
Example : free living
chimera_check - A chimeric sequence, or chimera for short, is a sequence comprised of two or more phylogenetically distinct parent sequences. Chimeras are usually PCR artifacts thought to occur when a prematurely terminated amplicon reanneals to a foreign DNA strand and is copied to completion in the following PCR cycles. The point at which the chimeric sequence changes from one parent to the next is called the breakpoint or conversion point .
Section : sequencing
Expected value : name and version of software, parameters used
Value syntax : {software};{version};{parameters}
Example : uchime;v4.1;default parameters
compl_appr - The approach used to determine the completeness of a given SAG or MAG, which would typically make use of a set of conserved marker genes or a closely related reference genome. For UViG completeness, include reference genome or group used, and contig feature suggesting a complete genome.
Section : sequencing
Expected value : enumeration
Value syntax : [marker gene|reference based|other]
Example : other: UViG length compared to the average length of reference genomes from the P22virus genus (NCBI RefSeq v83)
compl_score - Completeness score is typically based on either the fraction of markers found as compared to a database or the percent of a genome found as compared to a closely related reference genome. High Quality Draft: >90%, Medium Quality Draft: >50%, and Low Quality Draft: < 50% should have the indicated completeness scores.
Section : sequencing
Expected value : quality;percent completeness
Value syntax : [high|med|low];{percentage}
Example : med;60%
compl_software - Tools used for completion estimate, i.e. checkm, anvi’o, busco.
Section : sequencing
Expected value : names and versions of software(s) used
Value syntax : {software};{version}
Example : checkm
contam_score - The contamination score is based on the fraction of single-copy genes that are observed more than once in a query genome. The following scores are acceptable for; High Quality Draft: < 5%, Medium Quality Draft: < 10%, Low Quality Draft: < 10%. Contamination must be below 5% for a SAG or MAG to be deposited into any of the public databases.
Section : sequencing
Expected value : value
Value syntax : {float} percentage
Example : 0.01
contam_screen_input - The type of sequence data used as input.
Section : sequencing
Expected value : enumeration
Value syntax : [reads| contigs]
Example : contigs
contam_screen_param - Specific parameters used in the decontamination sofware, such as reference database, coverage, and kmers. Combinations of these parameters may also be used, i.e. kmer and coverage, or reference database and kmer.
Section : sequencing
Expected value : enumeration;value or name
Value syntax : [ref db|kmer|coverage|combination];{text|integer}
Example : kmer
decontam_software - Tool(s) used in contamination screening.
Section : sequencing
Expected value : enumeration
Value syntax : [checkm/refinem|anvi’o|prodege|bbtools:decontaminate.sh|acdc|combination]
Example : anvi’o
detec_type - Type of UViG detection.
Section : sequencing
Expected value : enumeration
Value syntax : [independent sequence (UViG)|provirus (UpViG)]
Example : independent sequence (UViG)
encoded_traits - Should include key traits like antibiotic resistance or xenobiotic degradation phenotypes for plasmids, converting genes for phage.
Section : nucleic acid sequence source
Expected value : for plasmid: antibiotic resistance; for phage: converting genes
Value syntax : {text}
Example : beta-lactamase class A
env_package - MIxS extension for reporting of measurements and observations obtained from one or more of the environments where the sample was obtained. All environmental packages listed here are further defined in separate subtables. By giving the name of the environmental package, a selection of fields can be made from the subtables and can be reported.
Section : mixs extension
Expected value : enumeration
Value syntax : [air|built environment|host-associated|human-associated|human-skin|human-oral|human-gut|human-vaginal|hydrocarbon resources-cores|hydrocarbon resources-fluids/swabs|microbial mat/biofilm|misc environment|plant-associated|sediment|soil|wastewater/sludge|water]
Example : soil
estimated_size - The estimated size of the genome prior to sequencing. Of particular importance in the sequencing of (eukaryotic) genome which could remain in draft form for a long or unspecified period..
Section : nucleic acid sequence source
Expected value : number of base pairs
Value syntax : {integer} bp
Example : 300000 bp
experimental_factor - Experimental factors are essentially the variable aspects of an experiment design which can be used to describe an experiment, or set of experiments, in an increasingly detailed manner. This field accepts ontology terms from Experimental Factor Ontology (EFO) and/or Ontology for Biomedical Investigations (OBI). For a browser of EFO (v 2.95) terms, please see http://purl.bioontology.org/ontology/EFO; for a browser of OBI (v 2018-02-12) terms please see http://purl.bioontology.org/ontology/OBI.
Section : investigation
Expected value : text or EFO and/or OBI
Value syntax : {termLabel} {[termID]}|{text}
Example : time series design [EFO:EFO_0001779]
extrachrom_elements - Do plasmids exist of significant phenotypic consequence (e.g. ones that determine virulence or antibiotic resistance). Megaplasmids? Other plasmids (borrelia has 15+ plasmids).
Section : nucleic acid sequence source
Expected value : number of extrachromosmal elements
Value syntax : {integer}
Example : 5
feat_pred - Method used to predict UViGs features such as ORFs, integration site, etc..
Section : sequencing
Expected value : names and versions of software(s), parameters used
Value syntax : {software};{version};{parameters}
Example : Prodigal;2.6.3;default parameters
health_disease_stat - Health or disease status of specific host at time of collection.
Section : nucleic acid sequence source
Expected value : enumeration
Value syntax : [healthy|diseased|dead|disease-free|undetermined|recovering|resolving|pre-existing condition|pathological|life threatening|congenital]
Example : dead
host_pred_appr - Tool or approach used for host prediction.
Section : sequencing
Expected value : enumeration
Value syntax : [provirus|host sequence similarity|CRISPR spacer match|kmer similarity|co-occurrence|combination|other]
Example : CRISPR spacer match
host_pred_est_acc - For each tool or approach used for host prediction, estimated false discovery rates should be included, either computed de novo or from the literature.
Section : sequencing
Expected value : false discovery rate
Value syntax : {text}
Example : CRISPR spacer match: 0 or 1 mismatches, estimated 8% FDR at the host genus rank (Edwards et al. 2016 doi:10.1093/femsre/fuv048)
host_spec_range - The NCBI taxonomy identifier of the specific host if it is known.
Section : nucleic acid sequence source
Expected value : NCBI taxid
Value syntax : {integer}
Example : 9606
isol_growth_condt - Publication reference in the form of pubmed ID (pmid), digital object identifier (doi) or url for isolation and growth condition specifications of the organism/material.
Section : nucleic acid sequence source
Expected value : PMID,DOI or URL
Value syntax : {PMID}|{DOI}|{URL}
Example : doi: 10.1016/j.syapm.2018.01.009
lib_layout - Specify whether to expect single, paired, or other configuration of reads.
Section : sequencing
Expected value : enumeration
Value syntax : [paired|single|vector|other]
Example : paired
lib_reads_seqd - Total number of clones sequenced from the library.
Section : sequencing
Expected value : number of reads sequenced
Value syntax : {integer}
Example : 20
lib_screen - Specific enrichment or screening methods applied before and/or after creating libraries.
Section : sequencing
Expected value : screening strategy name
Value syntax : {text}
Example : enriched, screened, normalized
lib_size - Total number of clones in the library prepared for the project.
Section : sequencing
Expected value : number of clones
Value syntax : {integer}
Example : 50
lib_vector - Cloning vector type(s) used in construction of libraries.
Section : sequencing
Expected value : vector
Value syntax : {text}
Example : Bacteriophage P1
mag_cov_software - Tool(s) used to determine the genome coverage if coverage is used as a binning parameter in the extraction of genomes from metagenomic datasets.
Section : sequencing
Expected value : enumeration
Value syntax : [bwa|bbmap|bowtie|other]
Example : bbmap
mid - Molecular barcodes, called Multiplex Identifiers (MIDs), that are used to specifically tag unique samples in a sequencing run. Sequence should be reported in uppercase letters.
Section : sequencing
Expected value : multiplex identifier sequence
Value syntax : {dna}
Example : GTGAATAT
misc_param - Any other measurement performed or parameter collected, that is not listed here.
Section : soil
Expected value : parameter name;measurement value
Value syntax : {text};{float} {unit}
Example : Bicarbonate ion concentration;2075 micromole per kilogram
nucl_acid_amp - A link to a literature reference, electronic resource or a standard operating procedure (SOP), that describes the enzymatic amplification (PCR, TMA, NASBA) of specific nucleic acids.
Section : sequencing
Expected value : PMID, DOI or URL
Value syntax : {PMID}|{DOI}|{URL}
Example : https://phylogenomics.me/protocols/16s-pcr-protocol/
nucl_acid_ext - A link to a literature reference, electronic resource or a standard operating procedure (SOP), that describes the material separation to recover the nucleic acid fraction from a sample.
Section : sequencing
Expected value : PMID, DOI or URL
Value syntax : {PMID}|{DOI}|{URL}
Example : https://mobio.com/media/wysiwyg/pdfs/protocols/12888.pdf
num_replicons - Reports the number of replicons in a nuclear genome of eukaryotes, in the genome of a bacterium or archaea or the number of segments in a segmented virus. Always applied to the haploid chromosome count of a eukaryote.
Section : nucleic acid sequence source
Expected value : for eukaryotes and bacteria: chromosomes (haploid count); for viruses: segments
Value syntax : {integer}
Example : 2
number_contig - Total number of contigs in the cleaned/submitted assembly that makes up a given genome, SAG, MAG, or UViG.
Section : sequencing
Expected value : value
Value syntax : {integer}
Example : 40
pathogenicity - To what is the entity pathogenic.
Section : nucleic acid sequence source
Expected value : names of organisms that the entity is pathogenic to
Value syntax : {text}
Example : human, animal, plant, fungi, bacteria
pcr_cond - Description of reaction conditions and components of PCR in the form of ‘initial denaturation:94degC_1.5min; annealing=…’.
Section : sequencing
Expected value : initial denaturation:degrees_minutes;annealing:degrees_minutes;elongation:degrees_minutes;final elongation:degrees_minutes;total cycles
Value syntax : initial denaturation:degrees_minutes;annealing:degrees_minutes;elongation:degrees_minutes;final elongation:degrees_minutes;total cycles
Example : initial denaturation:94_3;annealing:50_1;elongation:72_1.5;final elongation:72_10;35
pcr_primers - PCR primers that were used to amplify the sequence of the targeted gene, locus or subfragment. This field should contain all the primers used for a single PCR reaction if multiple forward or reverse primers are present in a single PCR reaction. The primer sequence should be reported in uppercase letters.
Section : sequencing
Expected value : FWD: forward primer sequence;REV:reverse primer sequence
Value syntax : FWD:{dna};REV:{dna}
Example : FWD:GTGCCAGCMGCCGCGGTAA;REV:GGACTACHVGGGTWTCTAAT
ph - Ph measurement of the sample, or liquid portion of sample, or aqueous phase of the fluid.
Section : soil
Expected value : measurement value
Value syntax : {float}
Example : 7.2
ploidy - The ploidy level of the genome (e.g. allopolyploid, haploid, diploid, triploid, tetraploid). It has implications for the downstream study of duplicated gene and regions of the genomes (and perhaps for difficulties in assembly). For terms, please select terms listed under class ploidy (PATO:001374) of Phenotypic Quality Ontology (PATO), and for a browser of PATO (v 2018-03-27) please refer to http://purl.bioontology.org/ontology/PATO.
Section : nucleic acid sequence source
Expected value : PATO
Value syntax : {termLabel} {[termID]}
Example : allopolyploidy [PATO:0001379]
pred_genome_struc - Expected structure of the viral genome.
Section : sequencing
Expected value : enumeration
Value syntax : [segmented|non-segmented|undetermined]
Example : non-segmented
pred_genome_type - Type of genome predicted for the UViG.
Section : sequencing
Expected value : enumeration
Value syntax : [DNA|dsDNA|ssDNA|RNA|dsRNA|ssRNA|ssRNA (+)|ssRNA (-)|mixed|uncharacterized]
Example : dsDNA
propagation - This field is specific to different taxa. For phages: lytic/lysogenic, for plasmids: incompatibility group, for eukaryotes: sexual/asexual (Note: there is the strong opinion to name phage propagation obligately lytic or temperate, therefore we also give this choice.
Section : nucleic acid sequence source
Expected value : for virus: lytic, lysogenic, temperate, obligately lytic; for plasmid: incompatibility group; for eukaryote: asexual, sexual
Value syntax : {text}
Example : lytic
reassembly_bin - Has an assembly been performed on a genome bin extracted from a metagenomic assembly?.
Section : sequencing
Expected value : boolean
Value syntax : {boolean}
Example : no
ref_biomaterial - Primary publication if isolated before genome publication; otherwise, primary genome report.
Section : nucleic acid sequence source
Expected value : PMID, DOI or URL
Value syntax : {PMID}|{DOI}|{URL}
Example : doi:10.1016/j.syapm.2018.01.009
ref_db - List of database(s) used for ORF annotation, along with version number and reference to website or publication.
Section : sequencing
Expected value : names, versions, and references of databases
Value syntax : {database};{version};{reference}
Example : pVOGs;5; http://dmk-brain.ecn.uiowa.edu/pVOGs/ Grazziotin et al. 2017 doi:10.1093/nar/gkw975
rel_to_oxygen - Is this organism an aerobe, anaerobe? Please note that aerobic and anaerobic are valid descriptors for microbial environments.
Section : nucleic acid sequence source
Expected value : enumeration
Value syntax : [aerobe|anaerobe|facultative|microaerophilic|microanaerobe|obligate aerobe|obligate anaerobe]
Example : aerobe
samp_collect_device - The method or device employed for collecting the sample.
Section : nucleic acid sequence source
Expected value : type name
Value syntax : {text}
Example : biopsy, niskin bottle, push core
samp_mat_process - Any processing applied to the sample during or after retrieving the sample from environment. This field accepts OBI, for a browser of OBI (v 2018-02-12) terms please see http://purl.bioontology.org/ontology/OBI.
Section : nucleic acid sequence source
Expected value : text or OBI
Value syntax : {text}|{termLabel} {[termID]}
Example : filtering of seawater, storing samples in ethanol
samp_size - Amount or size of sample (volume, mass or area) that was collected.
Section : nucleic acid sequence source
Expected value : measurement value
Preferred unit : millliter, gram, milligram, liter
Value syntax : {float} {unit}
Example : 5 liter
samp_vol_we_dna_ext - Volume (ml), weight (g) of processed sample, or surface area swabbed from sample for DNA extraction.
Section : soil
Expected value : measurement value
Preferred unit : millliter, gram, milligram, square centimeter
Value syntax : {float} {unit}
Example : 1500 milliliter
seq_quality_check - Indicate if the sequence has been called by automatic systems (none) or undergone a manual editing procedure (e.g. by inspecting the raw data or chromatograms). Applied only for sequences that are not submitted to SRA,ENA or DRA.
Section : sequencing
Expected value : none or manually edited
Value syntax : [none|manually edited]
Example : none
sim_search_meth - Tool used to compare ORFs with database, along with version and cutoffs used.
Section : sequencing
Expected value : names and versions of software(s), parameters used
Value syntax : {software};{version};{parameters}
Example : HMMER3;3.1b2;hmmsearch, cutoff of 50 on score
single_cell_lysis_appr - Method used to free DNA from interior of the cell(s) or particle(s).
Section : sequencing
Expected value : enumeration
Value syntax : [chemical|enzymatic|physical|combination]
Example : enzymatic
single_cell_lysis_prot - Name of the kit or standard protocol used for cell(s) or particle(s) lysis.
Section : sequencing
Expected value : kit, protocol name
Value syntax : {text}
Example : ambion single cell lysis kit
size_frac - Filtering pore size used in sample preparation.
Section : nucleic acid sequence source
Expected value : filter size value range
Value syntax : {float}-{float} {unit}
Example : 0-0.22 micrometer
sop - Standard operating procedures used in assembly and/or annotation of genomes, metagenomes or environmental sequences.
Section : sequencing
Expected value : reference to SOP
Value syntax : {PMID}|{DOI}|{URL}
Example : http://press.igsb.anl.gov/earthmicrobiome/protocols-and-standards/its/
sort_tech - Method used to sort/isolate cells or particles of interest.
Section : sequencing
Expected value : enumeration
Value syntax : [flow cytometric cell sorting|microfluidics|lazer-tweezing|optical manipulation|micromanipulation|other]
Example : optical manipulation
source_mat_id - A unique identifier assigned to a material sample (as defined by http://rs.tdwg.org/dwc/terms/materialSampleID, and as opposed to a particular digital record of a material sample) used for extracting nucleic acids, and subsequent sequencing. The identifier can refer either to the original material collected or to any derived sub-samples. The INSDC qualifiers /specimen_voucher, /bio_material, or /culture_collection may or may not share the same value as the source_mat_id field. For instance, the /specimen_voucher qualifier and source_mat_id may both contain ‘UAM:Herps:14’ , referring to both the specimen voucher and sampled tissue with the same identifier. However, the /culture_collection qualifier may refer to a value from an initial culture (e.g. ATCC:11775) while source_mat_id would refer to an identifier from some derived culture from which the nucleic acids were extracted (e.g. xatc123 or ark:/2154/R2)..
Section : nucleic acid sequence source
Expected value : for cultures of microorganisms: identifiers for two culture collections; for other material a unique arbitrary identifer
Value syntax : {text}
Example : MPI012345
source_uvig - Type of dataset from which the UViG was obtained.
Section : nucleic acid sequence source
Expected value : enumeration
Value syntax : [metagenome (not viral targeted)|viral fraction metagenome (virome)|sequence-targeted metagenome|metatranscriptome (not viral targeted)|viral fraction RNA metagenome (RNA virome)|sequence-targeted RNA metagenome|microbial single amplified genome (SAG)|viral single amplified genome (vSAG)|isolate microbial genome|other]
Example : viral fraction metagenome (virome)
specific_host - If there is a host involved, please provide its taxid (or environmental if not actually isolated from the dead or alive host - i.e. a pathogen could be isolated from a swipe of a bench etc) and report whether it is a laboratory or natural host).
Section : nucleic acid sequence source
Expected value : host taxid, unknown, environmental
Value syntax : {NCBI taxid}|{text}
Example : 9606
subspecf_gen_lin - This should provide further information about the genetic distinctness of the sequenced organism by recording additional information e.g. serovar, serotype, biotype, ecotype, or any relevant genetic typing schemes like Group I plasmid. It can also contain alternative taxonomic information. It should contain both the lineage name, and the lineage rank, i.e. biovar:abc123.
Section : nucleic acid sequence source
Expected value : genetic lineage below lowest rank of NCBI taxonomy, which is subspecies, e.g. serovar, biotype, ecotype
Value syntax : {rank name}:{text}
Example : serovar:Newport
target_gene - Targeted gene or locus name for marker gene studies.
Section : sequencing
Expected value : gene name
Value syntax : {text}
Example : 16S rRNA, 18S rRNA, nif, amoA, rpo
target_subfragment - Name of subfragment of a gene or locus. Important to e.g. identify special regions on marker genes like V6 on 16S rRNA.
Section : sequencing
Expected value : gene fragment name
Value syntax : {text}
Example : V6, V9, ITS
tax_class - Method used for taxonomic classification, along with reference database used, classification rank, and thresholds used to classify new genomes.
Section : sequencing
Expected value : classification method, database name, and other parameters
Value syntax : {text}
Example : vConTACT vContact2 (references from NCBI RefSeq v83, genus rank classification, default parameters)
tax_ident - The phylogenetic marker(s) used to assign an organism name to the SAG or MAG.
Section : sequencing
Expected value : enumeration
Value syntax : [16S rRNA gene|multi-marker approach|other]
Example : other: rpoB gene
tot_nitro_content - Total nitrogen content of the sample.
Section : soil
Expected value : measurement value
Preferred unit : microgram per liter, micromole per liter, milligram per liter
Value syntax : {float} {unit}
tot_org_carb - Definition for soil: total organic carbon content of the soil, definition otherwise: total organic carbon content.
Section : soil
Expected value : measurement value
Preferred unit : gram Carbon per kilogram sample material
Value syntax : {float} {unit}
trna_ext_software - Tools used for tRNA identification.
Section : sequencing
Expected value : names and versions of software(s), parameters used
Value syntax : {software};{version};{parameters}
Example : infernal;v2;default parameters
trnas - The total number of tRNAs identified from the SAG or MAG.
Section : sequencing
Expected value : value from 0-21
Value syntax : {integer}
Example : 18
trophic_level - Trophic levels are the feeding position in a food chain. Microbes can be a range of producers (e.g. chemolithotroph).
Section : nucleic acid sequence source
Expected value : enumeration
Value syntax : [autotroph|carboxydotroph|chemoautotroph|chemoheterotroph|chemolithoautotroph|chemolithotroph|chemoorganoheterotroph|chemoorganotroph|chemosynthetic|chemotroph|copiotroph|diazotroph|facultative|autotroph|heterotroph|lithoautotroph|lithoheterotroph|lithotroph|methanotroph|methylotroph|mixotroph|obligate|chemoautolithotroph|oligotroph|organoheterotroph|organotroph|photoautotroph|photoheterotroph|photolithoautotroph|photolithotroph|photosynthetic|phototroph]
Example : heterotroph
url.
Section : sequencing
Expected value : URL
Value syntax : {URL}
Example : http://www.earthmicrobiome.org/
vir_ident_software - Tool(s) used for the identification of UViG as a viral genome, software or protocol name including version number, parameters, and cutoffs used.
Section : sequencing
Expected value : software name, version and relevant parameters
Value syntax : {software};{version};{parameters}
Example : VirSorter; 1.0.4; Virome database, category 2
virus_enrich_appr - List of approaches used to enrich the sample for viruses, if any.
Section : nucleic acid sequence source
Expected value : enumeration
Value syntax : [filtration|ultrafiltration|centrifugation|ultracentrifugation|PEG Precipitation|FeCl Precipitation|CsCl density gradient|DNAse|RNAse|targeted sequence capture|other|none]
Example : filtration + FeCl Precipitation + ultracentrifugation + DNAse
votu_class_appr - Cutoffs and approach used when clustering new UViGs in “species-level” vOTUs. Note that results from standard 95% ANI / 85% AF clustering should be provided alongside vOTUS defined from another set of thresholds, even if the latter are the ones primarily used during the analysis.
Section : sequencing
Expected value : cutoffs and method used
Value syntax : {ANI cutoff};{AF cutoff};{clustering method}
Example : 95% ANI;85% AF; greedy incremental clustering
votu_db - Reference database (i.e. sequences not generated as part of the current study) used to cluster new genomes in “species-level” vOTUs, if any.
Section : sequencing
Expected value : database and version
Value syntax : {database};{version}
Example : NCBI Viral RefSeq;83
votu_seq_comp_appr - Tool and thresholds used to compare sequences when computing “species-level” vOTUs.
Section : sequencing
Expected value : software name, version and relevant parameters
Value syntax : {software};{version};{parameters}
Example : blastn;2.6.0+;e-value cutoff: 0.001
water_content - Water content measurement.
Section : soil
Expected value : measurement value
Preferred unit : gram per gram or cubic centimeter per cubic centimeter
Value syntax : {float}
wga_amp_appr - Method used to amplify genomic DNA in preparation for sequencing.
Section : sequencing
Expected value : enumeration
Value syntax : [pcr based|mda based]
Example : mda based
wga_amp_kit - Kit used to amplify genomic DNA in preparation for sequencing.
Section : sequencing
Expected value : kit name
Value syntax : {text}
Example : qiagen repli-g
Credits
this project was made using the LinkML framework