NMDC Workflow Documentations
Overview
NMDC
The National Microbiome Data Collaborative (NMDC) is a new initiative, launched in July 2019 and funded by the Department of Energy’s (DOE) Office of Science, Biological and Environmental Research program, that aims to empower the research community to more effectively harness microbiome data. The NMDC is building an open-source, integrated data science ecosystem aimed at leveraging existing efforts in the microbiome research space, including data standards, quality, integration, and access, to create a linked data discovery portal. Read the Nature Reviews Microbiology Comment on the NMDC or visit the NMDC website.
Four national laboratories are working together to produce the NMDC:
Lawrence Berkeley National Laboratory
Los Alamos National Laboratory
Oak Ridge National Laboratory
Pacific Northwest National Laboratory
NMDC Workflows
General Guidelines
NMDC aims to integrate existing open-source bioinformatics tools into standardized workflows for processing raw multi-omics data to produce interoperable and reusable annotated data products. Any commercial software are optional alternatives and not required.
Execution Evironment
Two common ways to install and run the NMDC workflows:
Native installation
Containers
The NMDC workflows have been written in WDL and require a WDL-capable Workflow Execution Tool (i.e., Cromwell). To ease the native installation, Docker images have been created for the third-party tools for all of the workflows as well. The workflows use the corresponding Docker images to run the required third-party tools. Databases must be downloaded and installed for most of the workflows.
The NMDC workflows are also available as a web application called NMDC EDGE . The application has only the NMDC workflows integrated into an updated framework for EDGE Bioinformatics ; this provides the workflows, third-party software, and requisite databases within a platform with a user-friendly interface. NMDC EDGE is provided as a web application especially for users who are not comfortable with running command line tools or without the computational resources to run the command line/ Docker versions.
Reads QC Workflow (v1.0.2)
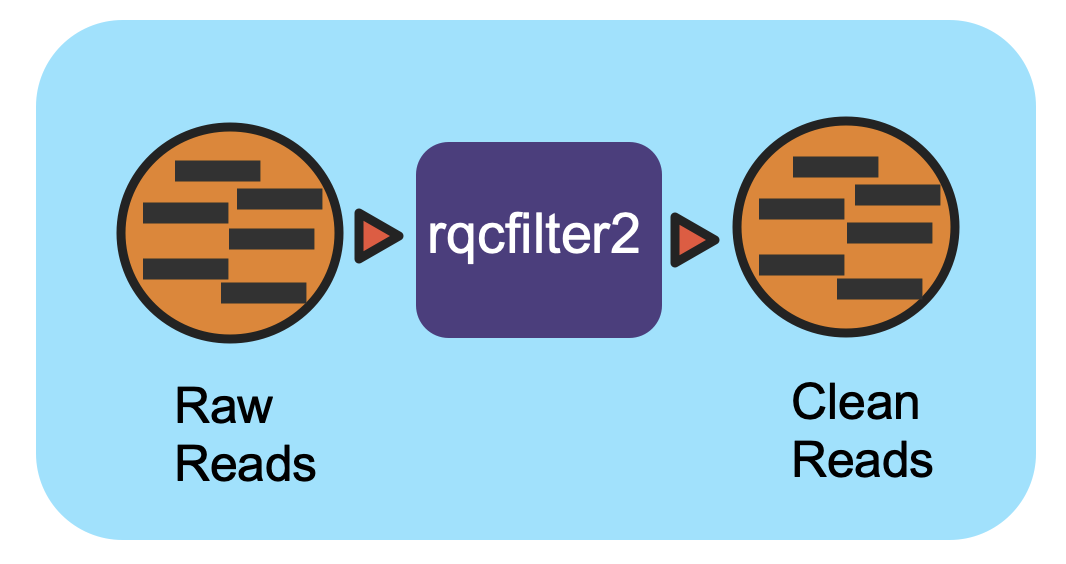
Workflow Overview
This workflow utilizes the program “rqcfilter2” from BBTools to perform quality control on raw Illumina reads. The workflow performs quality trimming, artifact removal, linker trimming, adapter trimming, and spike-in removal (using BBDuk), and performs human/cat/dog/mouse/microbe removal (using BBMap).
- The following parameters are used for “rqcfilter2” in this workflow::
qtrim=r : Quality-trim from right ends before mapping.
trimq=0 : Trim quality threshold.
maxns=3 : Reads with more Ns than this will be discarded.
maq=3 : Reads with average quality (before trimming) below this will be discarded.
minlen=51 : Reads shorter than this after trimming will be discarded. Pairs will be discarded only if both are shorter.
mlf=0.33 : Reads shorter than this fraction of original length after trimming will be discarded.
phix=true : Remove reads containing phiX kmers.
khist=true : Generate a kmer-frequency histogram of the output data.
kapa=true : Remove and quantify kapa tag
trimpolyg=5 : Trim reads that start or end with a G polymer at least this long
clumpify=true : Run clumpify; all deduplication flags require this.
removehuman=true : Remove human reads via mapping.
removedog=true : Remove dog reads via mapping.
removecat=true : Remove cat reads via mapping.
removemouse=true : Remove mouse reads via mapping.
barcodefilter=false : Disable improper barcodes filter
chastityfilter=false: Remove illumina reads failing chastity filter.
trimfragadapter=true: Trim all known Illumina adapter sequences, including TruSeq and Nextera.
removemicrobes=true : Remove common contaminant microbial reads via mapping, and place them in a separate file.
Workflow Availability
The workflow from GitHub uses all the listed docker images to run all third-party tools. The workflow is available in GitHub: https://github.com/microbiomedata/ReadsQC; the corresponding Docker image is available in DockerHub: https://hub.docker.com/r/microbiomedata/bbtools.
Requirements for Execution
(recommendations are in bold)
WDL-capable Workflow Execution Tool (Cromwell)
Container Runtime that can load Docker images (Docker v2.1.0.3 or higher)
Hardware Requirements
Disk space: 106 GB for the RQCFilterData database
Memory: >40 GB RAM
Workflow Dependencies
Third party software (This is included in the Docker image.)
BBTools v38.96 (License: BSD-3-Clause-LBNL)
Requisite database
The RQCFilterData Database must be downloaded and installed. This is a 106 GB tar file which includes reference datasets of artifacts, adapters, contaminants, the phiX genome, and some host genomes.
The following commands will download the database:
mkdir refdata
wget http://portal.nersc.gov/dna/microbial/assembly/bushnell/RQCFilterData.tar
tar -xvf RQCFilterData.tar -C refdata
rm RQCFilterData.tar
Sample dataset(s)
small dataset: Ecoli 10x . You can find input/output in the downloaded tar gz file.
large dataset: Zymobiomics mock-community DNA control (SRR7877884); the original gzipped dataset is ~5.7 GB. You can find input/output in the downloaded tar gz file.
Note
If the input data is paired-end data, it must be in interleaved format. The following command will interleave the files, using the above dataset as an example:
paste <(zcat SRR7877884_1.fastq.gz | paste - - - -) <(zcat SRR7877884_2.fastq.gz | paste - - - -) | tr '\t' '\n' | gzip -c > SRR7877884-int.fastq.gz
For testing purposes and for the following examples, we used a 10% sub-sampling of the above dataset: SRR7877884-int-0.1.fastq.gz. This dataset is already interleaved.
Inputs
A JSON file containing the following information:
the path to the database
the path to the interleaved fastq file (input data)
the path to the output directory
input_interleaved (boolean)
forwards reads fastq file (when input_interleaved is false)
reverse reads fastq file (when input_interleaved is false)
(optional) parameters for memory
(optional) number of threads requested
An example input JSON file is shown below:
{
"jgi_rqcfilter.database": "/path/to/refdata",
"jgi_rqcfilter.input_files": [
"/path/to/SRR7877884-int-0.1.fastq.gz "
],
"jgi_rqcfilter.input_interleaved": true,
"jgi_rqcfilter.input_fq1":[],
"jgi_rqcfilter.input_fq2":[],
"jgi_rqcfilter.outdir": "/path/to/rqcfiltered",
"jgi_rqcfilter.memory": "35G",
"jgi_rqcfilter.threads": "16"
}
Note
In an HPC environment, parallel processing allows for processing multiple samples. The “jgi_rqcfilter.input_files” parameter is an array data structure. It can be used for multiple samples as input separated by a comma (,). Ex: “jgi_rqcfilter.input_files”:[“first-int.fastq”,”second-int.fastq”]
Output
A directory named with the prefix of the FASTQ input file will be created and multiple output files are generated; the main QC FASTQ output is named prefix.anqdpht.fastq.gz. Using the dataset above as an example, the main output would be named SRR7877884-int-0.1.anqdpht.fastq.gz. Other files include statistics on the quality of the data; what was trimmed, detected, and filtered in the data; a status log, and a shell script documenting the steps implemented so the workflow can be reproduced.
An example output JSON file (filterStats.json) is shown below:
{
"inputReads": 331126,
"kfilteredBases": 138732,
"qfilteredReads": 0,
"ktrimmedReads": 478,
"outputBases": 1680724,
"ktrimmedBases": 25248,
"kfilteredReads": 926,
"qtrimmedBases": 0,
"outputReads": 11212,
"gcPolymerRatio": 0.182857,
"inputBases": 50000026,
"qtrimmedReads": 0,
"qfilteredBases": 0
}
Below is an example of all the output directory files with descriptions to the right.
FileName |
Description |
|---|---|
SRR7877884-int-0.1.anqdpht.fastq.gz |
main output (clean data) |
adaptersDetected.fa |
adapters detected and removed |
bhist.txt |
base composition histogram by position |
cardinality.txt |
estimation of the number of unique kmers |
commonMicrobes.txt |
detected common microbes |
file-list.txt |
output file list for rqcfilter2.sh |
filterStats.txt |
summary statistics |
filterStats.json |
summary statistics in JSON format |
filterStats2.txt |
more detailed summary statistics |
gchist.txt |
GC content histogram |
human.fq.gz |
detected human sequence reads |
ihist_merge.txt |
insert size histogram |
khist.txt |
kmer-frequency histogram |
kmerStats1.txt |
synthetic molecule (phix, linker, lamda, pJET) filter run log |
kmerStats2.txt |
synthetic molecule (short contamination) filter run log |
ktrim_kmerStats1.txt |
detected adapters filter run log |
ktrim_scaffoldStats1.txt |
detected adapters filter statistics |
microbes.fq.gz |
detected common microbes sequence reads |
microbesUsed.txt |
common microbes list for detection |
peaks.txt |
number of unique kmers in each peak on the histogram |
phist.txt |
polymer length histogram |
refStats.txt |
human reads filter statistics |
reproduce.sh |
the shell script to reproduce the run |
scaffoldStats1.txt |
detected synthetic molecule (phix, linker, lamda, pJET) statistics |
scaffoldStats2.txt |
detected synthetic molecule (short contamination) statistics |
scaffoldStatsSpikein.txt |
detected skipe-in kapa tag statistics |
sketch.txt |
mash type sketch scanned result against nt, refseq, silva database sketches. |
spikein.fq.gz |
detected skipe-in kapa tag sequence reads |
status.log |
rqcfilter2.sh running log |
synth1.fq.gz |
detected synthetic molecule (phix, linker, lamda, pJET) sequence reads |
synth2.fq.gz |
detected synthetic molecule (short contamination) sequence reads |
Version History
1.0.2 (release date 04/09/2021; previous versions: 1.0.1)
Point of contact
Original author: Brian Bushnell <bbushnell@lbl.gov>
Package maintainer: Chienchi Lo <chienchi@lanl.gov>
The Read-based Taxonomy Classification (v1.0.1)
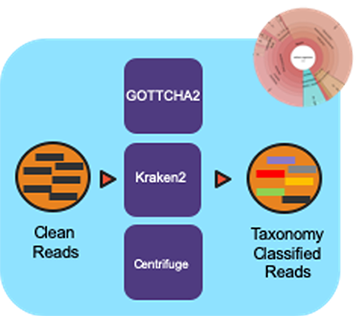
Workflow Overview
The pipeline takes in sequencing files (single- or paired-end) and profiles them using multiple taxonomic classification tools with the Cromwell as the workflow manager.
Workflow Availability
The workflow is available in GitHub: https://github.com/microbiomedata/ReadbasedAnalysis; the corresponding Docker image is available in DockerHub: https://hub.docker.com/r/microbiomedata/nmdc_taxa_profilers
Requirements for Execution:
(recommendations are in bold)
WDL-capable Workflow Execution Tool (Cromwell)
Container Runtime that can load Docker images (Docker v2.1.0.3 or higher)
Hardware Requirements:
Disk space: 152 GB for databases (55 GB, 89 GB, and 8 GB for GOTTCHA2, Kraken2 and Centrifuge databases, respectively)
60 GB RAM
Workflow Dependencies
Third party software:
(These are included in the Docker image.)
GOTTCHA2 v2.1.6 (License: BSD-3-Clause-LANL)
Kraken2 v2.0.8 (License: MIT)
Centrifuge v1.0.4 (License: GPL-3)
Requisite databases:
The database for each tool must be downloaded and installed. These databases total 152 GB.
GOTTCHA2 database (gottcha2/):
The database RefSeqr90.cg.BacteriaArchaeaViruses.species.fna contains complete genomes of bacteria, archaea and viruses from RefSeq Release 90. The following commands will download the database:
wget https://edge-dl.lanl.gov/GOTTCHA2/RefSeq-r90.cg.BacteriaArchaeaViruses.species.tar
tar -xvf RefSeq-r90.cg.BacteriaArchaeaViruses.species.tar
rm RefSeq-r90.cg.BacteriaArchaeaViruses.species.tar
Kraken2 database (kraken2/):
This is a standard Kraken 2 database, built from NCBI RefSeq genomes. The following commands will download the database:
mkdir kraken2
wget https://genome-idx.s3.amazonaws.com/kraken/k2_standard_20201202.tar.gz
tar -xzvf k2_standard_20201202.tar.gz -C kraken2
rm k2_standard_20201202.tar.gz
Centrifuge database (centrifuge/):
This is a compressed database built from RefSeq genomes of Bacteria and Archaea. The following commands will download the database:
mkdir centrifuge
wget https://genome-idx.s3.amazonaws.com/centrifuge/p_compressed_2018_4_15.tar.gz
tar -xzvf p_compressed_2018_4_15.tar.gz -C centrifuge
rm p_compressed_2018_4_15.tar.gz
Sample dataset(s):
Zymobiomics mock-community DNA control (SRR7877884); this dataset is ~7 GB.
Input: A JSON file containing the following information: 1. selection of profiling tools (set as true if selected) 2. the paths to the required database(s) for the tools selected 3. the paths to the input fastq file(s) (paired-end data is shown; this can be the output of the Reads QC workflow in interleaved format which will be treated as single-end data.) 4. the prefix for the output file names 5. the path of the output directory 6. CPU number requested for the run.
{
"ReadbasedAnalysis.enabled_tools": {
"gottcha2": true,
"kraken2": true,
"centrifuge": true
},
"ReadbasedAnalysis.db": {
"gottcha2": "/path/to/database/RefSeq-r90.cg.BacteriaArchaeaViruses.species.fna",
"kraken2": " /path/to/kraken2",
"centrifuge": "/path/to/centrifuge/p_compressed"
},
"ReadbasedAnalysis.reads": [
"/path/to/SRR7877884.1.fastq.gz",
"/path/to/SRR7877884.2.fastq.gz"
],
"ReadbasedAnalysis.paired": true,
"ReadbasedAnalysis.prefix": "SRR7877884",
"ReadbasedAnalysis.outdir": "/path/to/ReadbasedAnalysis",
"ReadbasedAnalysis.cpu": 4
}
Output:
The workflow creates an output JSON file and individual output sub-directories for each tool which include tabular classification results, a tabular report, and a Krona plot (html).:
ReadbasedAnalysis/
|-- SRR7877884.json
|-- centrifuge
| |-- SRR7877884.classification.tsv
| |-- SRR7877884.report.tsv
| `-- SRR7877884.krona.html
|
|-- gottcha2
| |-- SRR7877884.full.tsv
| |-- SRR7877884.krona.html
| `-- SRR7877884.tsv
|
`-- kraken2
|-- SRR7877884.classification.tsv
|-- SRR7877884.krona.html
`-- SRR7877884.report.tsv
Below is an example of the output directory files with descriptions to the right.
FileName |
Description |
SRR7877884.json |
ReadbasedAnalysis result JSON file |
centrifuge/SRR7877884.classification.tsv |
Centrifuge output read classification TSV file |
centrifuge/SRR7877884.report.tsv |
Centrifuge output report TSV file |
centrifuge/SRR7877884.krona.html |
Centrifuge krona plot HTML file |
gottcha2/SRR7877884.full.tsv |
GOTTCHA2 detail output TSV file |
gottcha2/SRR7877884.tsv |
GOTTCHA2 output report TSV file |
gottcha2/SRR7877884.krona.html |
GOTTCHA2 krona plot HTML file |
kraken2/SRR7877884.classification.tsv |
Kraken2 output read classification TSV file |
kraken2/SRR7877884.report.tsv |
Kraken2 output report TSV file |
kraken2/SRR7877884.krona.html |
Kraken2 krona plot HTML file |
Version History
1.0.1 (release date 01/14/2021; previous versions: 1.0.0)
Point of contact
Package maintainer: Po-E Li <po-e@lanl.gov>
Metagenome Assembly Workflow (v1.0.2)
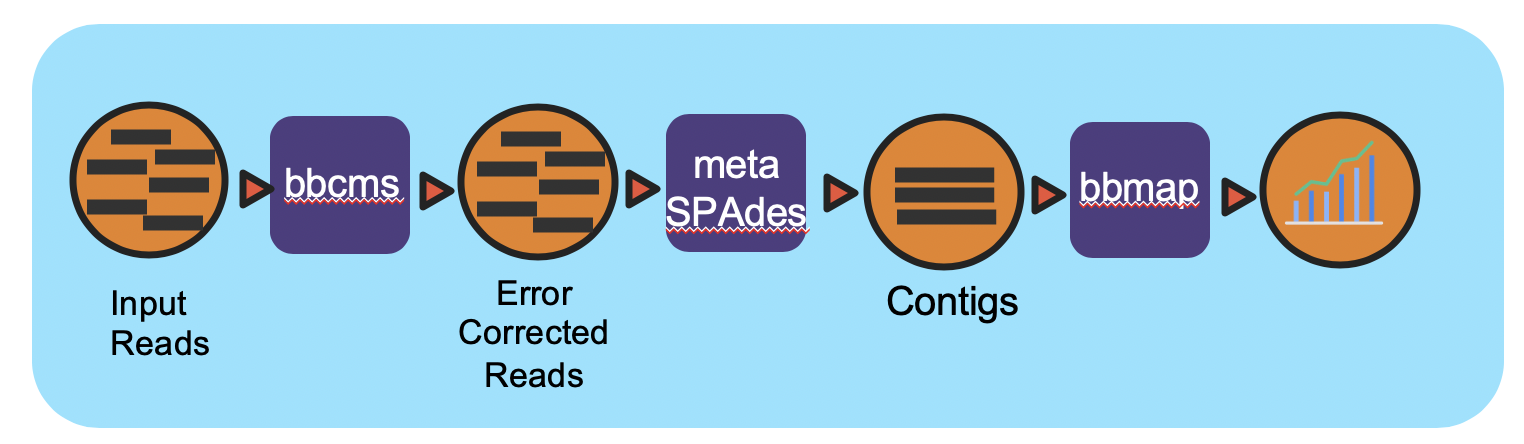
Workflow Overview
This workflow takes in paired-end Illumina reads in interleaved format and performs error correction, then reformats the interleaved file into two FASTQ files for downstream tasks using bbcms (BBTools). The corrected reads are assembled using metaSPAdes. After assembly, the reads are mapped back to contigs by bbmap (BBTools) for coverage information. The .wdl (Workflow Description Language) file includes five tasks, bbcms, assy, create_agp, read_mapping_pairs, and make_output.
The bbcms task takes in interleaved FASTQ inputs and performs error correction and reformats the interleaved fastq into two output FASTQ files for paired-end reads for the next tasks.
The assy task performs metaSPAdes assembly
Contigs and Scaffolds (output of metaSPAdes) are consumed by the create_agp task to rename the FASTA header and generate an AGP format which describes the assembly
The read_mapping_pairs task maps reads back to the final assembly to generate coverage information.
The final make_output task adds all output files into the specified directory.
Workflow Availability
The workflow from GitHub uses all the listed docker images to run all third-party tools. The workflow is available in GitHub: https://github.com/microbiomedata/metaAssembly; the corresponding Docker images are available in DockerHub: https://hub.docker.com/r/microbiomedata/spades and https://hub.docker.com/r/microbiomedata/bbtools
Requirements for Execution
(recommendations are in bold)
WDL-capable Workflow Execution Tool (Cromwell)
Container Runtime that can load Docker images (Docker v2.1.0.3 or higher)
Hardware Requirements
Memory: >40 GB RAM
The memory requirement depends on the input complexity. Here is a simple estimation equation for the memory required based on kmers in the input file:
predicted_mem = (kmers * 2.962e-08 + 1.630e+01) * 1.1 (in GB)
Note
The kmers variable for the equation above can be obtained using the kmercountmulti.sh script from BBTools.
kmercountmulti.sh -k=31 in=your.read.fq.gz
Workflow Dependencies
Third party software: (This is included in the Docker image.)
metaSPades v3.15.0 (License: GPLv2)
BBTools:38.94 (License: BSD-3-Clause-LBNL)
Sample dataset(s)
small dataset: Ecoli 10x (287M) . You can find input/output in the downloaded tar gz file.
large dataset: Zymobiomics mock-community DNA control (22G) . You can find input/output in the downloaded tar gz file.
Zymobiomics mock-community DNA control (SRR7877884); this original dataset is ~4 GB.
For testing purposes and for the following examples, we used a 10% sub-sampling of the above dataset: (SRR7877884-int-0.1.fastq.gz). This dataset is already interleaved.
Input
A JSON file containing the following information:
the path to the input FASTQ file (Illumina paired-end interleaved FASTQ) (recommended the output of the Reads QC workflow.)
the contig prefix for the FASTA header
the output path
input_interleaved (boolean)
forwards reads fastq file (required value when input_interleaved is false, otherwise use [] )
reverse reads fastq file (required value when input_interleaved is false, otherwise use [] )
memory (optional) ex: “jgi_metaASM.memory”: “105G”
threads (optional) ex: “jgi_metaASM.threads”: “16”
An example input JSON file is shown below:
{
"jgi_metaASM.input_file":["/path/to/SRR7877884-int-0.1.fastq.gz "],
"jgi_metaASM.rename_contig_prefix":"projectID",
"jgi_metaASM.outdir":"/path/to/ SRR7877884-int-0.1_assembly",
"jgi_metaASM.input_interleaved":true,
"jgi_metaASM.input_fq1":[],
"jgi_metaASM.input_fq2":[],
"jgi_metaASM.memory": "105G",
"jgi_metaASM.threads": "16"
}
Output
The output directory will contain following files:
output/
├── assembly.agp
├── assembly_contigs.fna
├── assembly_scaffolds.fna
├── covstats.txt
├── pairedMapped.sam.gz
├── pairedMapped_sorted.bam
└── stats.json
Part of an example output stats JSON file is shown below:
``` {
“scaffolds”: 58, “contigs”: 58, “scaf_bp”: 28406, “contig_bp”: 28406, “gap_pct”: 0.00000, “scaf_N50”: 21, “scaf_L50”: 536, “ctg_N50”: 21, “ctg_L50”: 536, “scaf_N90”: 49, “scaf_L90”: 317, “ctg_N90”: 49, “ctg_L90”: 317, “scaf_logsum”: 22.158, “scaf_powsum”: 2.245, “ctg_logsum”: 22.158, “ctg_powsum”: 2.245, “asm_score”: 0.000, “scaf_max”: 1117, “ctg_max”: 1117, “scaf_n_gt50K”: 0, “scaf_l_gt50K”: 0, “scaf_pct_gt50K”: 0.0, “gc_avg”: 0.39129, “gc_std”: 0.03033, “filename”: “/global/cfs/cdirs/m3408/aim2/metagenome/assembly/cromwell-executions/jgi_metaASM/3342a6e8-7f78-40e6-a831-364dd2a47baa/call-create_agp/execution/assembly_scaffolds.fna”
The table provides all of the output directories, files, and their descriptions.
Directory |
File Name |
Description |
|---|---|---|
bbcms |
Error correction result directory |
|
bbcms/berkeleylab-jgi-meta-60ade422cd4e |
directory containing checking resource script |
|
bbcms/ |
counts.metadata.json |
bbcms commands and summary statistics in JSON format |
bbcms/ |
input.corr.fastq.gz |
error corrected reads in interleaved format. |
bbcms/ |
input.corr.left.fastq.gz |
error corrected forward reads |
bbcms/ |
input.corr.right.fastq.gz |
error corrected reverse reads |
bbcms/ |
rc |
cromwell script sbumit return code |
bbcms/ |
readlen.txt |
error corrected reads statistics |
bbcms/ |
resources.log |
resource checking log |
bbcms/ |
script |
Task run commands |
bbcms/ |
script.background |
Bash script to run script.submit |
bbcms/ |
script.submit |
cromwell submit commands |
bbcms/ |
stderr |
standard error where task writes error message to |
bbcms/ |
stderr.background |
standard error where bash script writes error message to |
bbcms/ |
stderr.log |
standard error from bbcms command |
bbcms/ |
stdout |
standard output where task writes error message to |
bbcms/ |
stdout.background |
standard output where bash script writes error message(s) |
bbcms/ |
stdout.log |
standard output from bbcms command |
bbcms/ |
unique31mer.txt |
the count of unique kmer, K=31 |
spades3 |
metaSPAdes assembly result directory |
|
spades3/K33 |
directory containing intermediate files from the run with K=33 |
|
spades3/K55 |
directory containing intermediate files from the run with K=55 |
|
spades3/K77 |
directory containing intermediate files from the run with K=77 |
|
spades3/K99 |
directory containing intermediate files from the run with K=99 |
|
spades3/K127 |
directory containing intermediate files from the run with K=127 |
|
spades3/misc |
directory containing miscellaneous files |
|
spades3/tmp |
directory for temp files |
|
spades3/ |
assembly_graph.fastg |
metaSPAdes assembly graph in FASTG format |
spades3/ |
assembly_graph_with_scaffolds.gfa |
metaSPAdes assembly graph and scaffolds paths in GFA 1.0 format |
spades3/ |
before_rr.fasta |
contigs before repeat resolution |
spades3/ |
contigs.fasta |
metaSPAdes resulting contigs |
spades3/ |
contigs.paths |
paths in the assembly graph corresponding to contigs.fasta |
spades3/ |
dataset.info |
internal configuration file |
spades3/ |
first_pe_contigs.fasta |
preliminary contigs of iterative kmers assembly |
spades3/ |
input_dataset.yaml |
internal YAML data set file |
spades3/ |
params.txt |
information about SPAdes parameters in this run |
spades3/ |
scaffolds.fasta |
metaSPAdes resulting scaffolds |
spades3/ |
scaffolds.paths |
paths in the assembly graph corresponding to scaffolds.fasta |
spades3/ |
spades.log |
metaSPAdes log |
final_assembly |
create_agp task result directory |
|
final_assembly/berkeleylab-jgi-meta-60ade422cd4e |
directory containing checking resource script |
|
final_assembly/ |
assembly.agp |
an AGP format file describes the assembly |
final_assembly/ |
assembly_contigs.fna |
Final assembly contig fasta |
final_assembly/ |
assembly_scaffolds.fna |
Final assembly scaffolds fasta |
final_assembly/ |
assembly_scaffolds.legend |
name mapping file from spades node name to new name |
final_assembly/ |
rc |
cromwell script sbumit return code |
final_assembly/ |
resources.log |
resource checking log |
final_assembly/ |
script |
Task run commands |
final_assembly/ |
script.background |
Bash script to run script.submit |
final_assembly/ |
script.submit |
cromwell submit commands |
final_assembly/ |
stats.json |
assembly statistics in json format |
final_assembly/ |
stderr |
standard error where task writes error message to |
final_assembly/ |
stderr.background |
standard error where bash script writes error message to |
final_assembly/ |
stdout |
standard output where task writes error message to |
final_assembly/ |
stdout.background |
standard output where bash script writes error message to |
mapping |
maps reads back to the final assembly result directory |
|
mapping/ |
covstats.txt |
contigs coverage informaiton |
mapping/ |
mapping_stats.txt |
contigs coverage informaiton (same as covstats.txt) |
mapping/ |
pairedMapped.bam |
reads mapping back to the final assembly bam file |
mapping/ |
pairedMapped.sam.gz |
reads mapping back to the final assembly sam.gz file |
mapping/ |
pairedMapped_sorted.bam |
reads mapping back to the final assembly sorted bam file |
mapping/ |
pairedMapped_sorted.bam.bai |
reads mapping back to the final assembly sorted bam index file |
mapping/ |
rc |
cromwell script sbumit return code |
mapping/ |
resources.log |
resource checking log |
mapping/ |
script |
Task run commands |
mapping/ |
script.background |
Bash script to run script.submit |
mapping/ |
script.submit |
cromwell submit commands |
mapping/ |
stderr |
standard error where task writes error message to |
mapping/ |
stderr.background |
standard error where bash script writes error message to |
mapping/ |
stdout |
standard output where task writes error message to |
mapping/ |
stdout.background |
standard output where bash script writes error message to |
Version History
1.0.2 (release date 03/12/2021; previous versions: 1.0.1)
Point of contact
Original author: Brian Foster <bfoster@lbl.gov>
Package maintainer: Chienchi Lo <chienchi@lanl.gov>
Metagenome Annotation Workflow (v1.0.0)
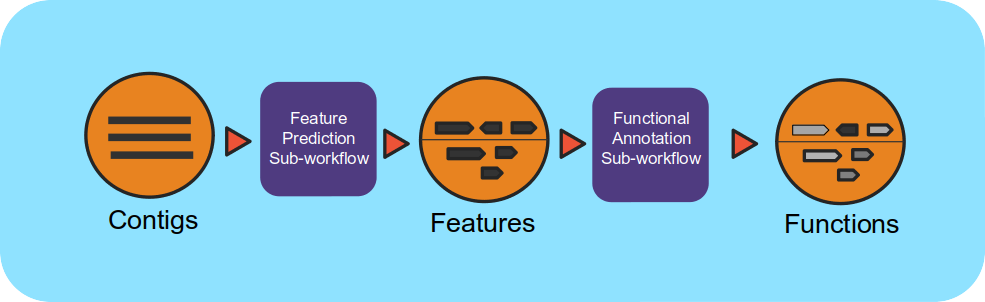
Workflow Overview
This workflow takes assembled metagenomes and generates structural and functional annotations. The workflow uses a number of open-source tools and databases to generate the structural and functional annotations.
The input assembly is first split into 10MB splits to be processed in parallel. Depending on the workflow engine configuration, the split can be processed in parallel. Each split is first structurally annotated, then those results are used for the functional annotation. The structural annotation uses tRNAscan_se, RFAM, CRT, Prodigal and GeneMarkS. These results are merged to create a consensus structural annotation. The resulting GFF is the input for functional annotation which uses multiple protein family databases (SMART, COG, TIGRFAM, SUPERFAMILY, Pfam and Cath-FunFam) along with custom HMM models. The functional predictions are created using Last and HMM. These annotations are also merged into a consensus GFF file. Finally, the respective split annotations are merged together to generate a single structural annotation file and single functional annotation file. In addition, several summary files are generated in TSV format.
Workflow Availability
The workflow is available in GitHub: https://github.com/microbiomedata/mg_annotation/ and the corresponding Docker image is available in DockerHub: https://hub.docker.com/r/microbiomedata/mg-annotation.
Requirements for Execution (recommendations are in bold):
WDL-capable Workflow Execution Tool (Cromwell)
Container Runtime that can load Docker images (Docker v2.1.0.3 or higher)
Hardware Requirements:
Disk space: 106 GB for the reference databases
Memory: >100 GB RAM
Workflow Dependencies
- Third party software (This is included in the Docker image.)
Conda (3-clause BSD)
tRNAscan-SE >= 2.0 (GNU GPL v3)
Infernal 1.1.2 (BSD)
CRT-CLI 1.8 (Public domain software, last official version is 1.2)
Prodigal 2.6.3 (GNU GPL v3)
GeneMarkS-2 >= 1.07 (Academic license for GeneMark family software)
Last >= 983 (GNU GPL v3)
HMMER 3.1b2 (3-clause BSD)
TMHMM 2.0 (Academic)
Requisite databases: The databases are available by request. Please contact NMDC (support@microbiomedata.org) for access.
Sample datasets
https://raw.githubusercontent.com/microbiomedata/mg_annotation/master/example.fasta
Input: A JSON file containing the following:
The path to the assembled contigs fasta file
The ID to associate with the result products (e.g. sample ID)
An example JSON file is shown below:
{
"annotation.imgap_input_fasta": "/path/to/fasta.fna",
"annotation.imgap_project_id": "samp_xyz123"}
}
Output: The final structural and functional annotation files are in GFF format and the summary files are in TSV format. The key outputs are listed below but additional files are available.
GFF: Structural annotation
GFF: Functional annotation
TSV: KO Summary
TSV: EC Summary
TSV: Gene Phylogeny Summar
The output paths can be obtained from the output metadata file from the Cromwell Exectuion. Here is a snippet from the outputs section of the full metadata JSON file.
{
"annotation.cath_funfam_gff": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_cath_funfam.gff",
"annotation.cog_gff": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_cog.gff",
"annotation.ko_ec_gff": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_ko_ec.gff",
"annotation.product_names_tsv": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_product_names.tsv",
"annotation.gene_phylogeny_tsv": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_gene_phylogeny.tsv",
"annotation.pfam_gff": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_pfam.gff",
"annotation.proteins_tigrfam_domtblout": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_proteins.tigrfam.domtblout",
"annotation.structural_gff": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_structural_annotation.gff",
"annotation.ec_tsv": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_ec.tsv",
"annotation.supfam_gff": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_supfam.gff",
"annotation.proteins_supfam_domtblout": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_proteins.supfam.domtblout",
"annotation.tigrfam_gff": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_tigrfam.gff",
"annotation.stats_tsv": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-final_stats/execution/samp_xyz123_structural_annotation_stats.tsv",
"annotation.proteins_cog_domtblout": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_proteins.cog.domtblout",
"annotation.ko_tsv": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_ko.tsv",
"annotation.proteins_pfam_domtblout": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_proteins.pfam.domtblout",
"annotation.proteins_smart_domtblout": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_proteins.smart.domtblout",
"annotation.crt_crisprs": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_crt.crisprs",
"annotation.functional_gff": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_functional_annotation.gff",
"annotation.proteins_faa": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123.faa",
"annotation.smart_gff": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_smart.gff",
"annotation.proteins_cath_funfam_domtblout": "/output/cromwell-executions/annotation/a67a5a0f-1ad7-4469-bb0c-780f4ef20307/call-merge_outputs/execution/samp_xyz123_proteins.cath_funfam.domtblout"
}
Version History: 1.0.0 (release data)
Point of contact
Package maintainer: Shane Canon <scanon@lbl.gov>
Metagenome Assembled Genomes Workflow (v1.0.4)
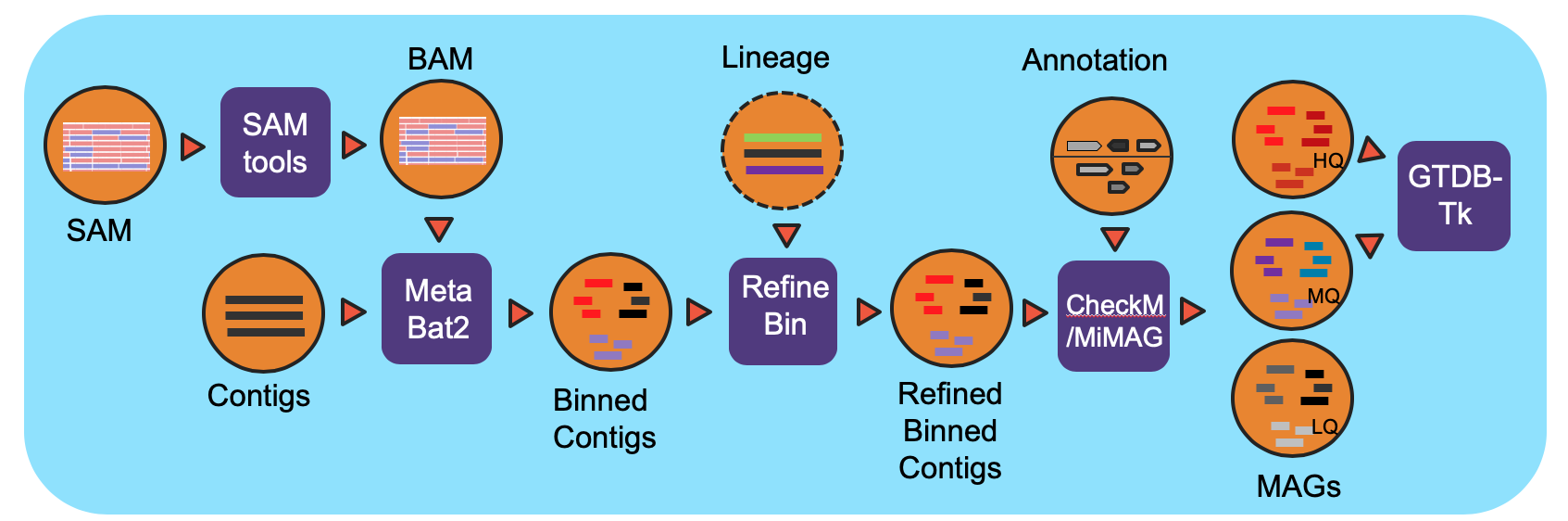
Workflow Overview
The workflow is based on IMG metagenome binning pipeline and has been modified specifically for the NMDC project. For all processed metagenomes, it classifies contigs into bins using MetaBat2. Next, the bins are refined using the functional Annotation file (GFF) from the Metagenome Annotation workflow and optional contig lineage information. The completeness of and the contamination present in the bins are evaluated by CheckM and bins are assigned a quality level (High Quality (HQ), Medium Quality (MQ), Low Quality (LQ)) based on MiMAG standards. In the end, GTDB-Tk is used to assign lineage for HQ and MQ bins.
Workflow Availability
The workflow from GitHub uses all the listed docker images to run all third-party tools. The workflow is available in GitHub: https://github.com/microbiomedata/metaMAGs The corresponding Docker image is available in DockerHub: https://hub.docker.com/r/microbiomedata/nmdc_mbin
Requirements for Execution
(recommendations are in bold):
WDL-capable Workflow Execution Tool (Cromwell)
Container Runtime that can load Docker images (Docker v2.1.0.3 or higher)
Hardware Requirements
Disk space: > 33 GB for the CheckM and GTDB-Tk databases
Memory: ~150GB memory for GTDB-tk.
Workflow Dependencies
Third party software (These are included in the Docker image.)
Biopython v1.74 (License: BSD-3-Clause)
Sqlite (License: Public Domain)
Pymysql (License: MIT License)
requests (License: Apache 2.0)
samtools > v1.9 (License: MIT License)
Metabat2 v2.15 (License: BSD-3-Clause)
CheckM v1.1.2 (License: GPLv3)
GTDB-TK v1.3.0 (License: GPLv3)
FastANI v1.3 (License: Apache 2.0)
FastTree v2.1.10 (License: GPLv2)
Requisite databases
The GTDB-Tk database must be downloaded and installed. The CheckM database included in the Docker image is a 275MB file contains the databases used for the Metagenome Binned contig quality assessment. The GTDB-Tk (27GB) database is used to assign lineages to the binned contigs.
The following commands will download and unarchive the GTDB-Tk database:
wget https://data.gtdb.ecogenomic.org/releases/release95/95.0/auxillary_files/gtdbtk_r95_data.tar.gz tar -xvzf gtdbtk_r95_data.tar.gz mv release95 GTDBTK_DB rm gtdbtk_r95_data.tar.gz
Sample dataset(s)
The following test datasets include an assembled contigs file, a BAM file, and a functional annotation file:
small dataset: LQ only (3.1G) . You can find input/output in the downloaded tar gz file.
large dataset: with HQ and MQ bins (12G) . You can find input/output in the downloaded tar gz file.
Input
A JSON file containing the following:
the number of CPUs requested
The number of threads used by pplacer (Use lower number to reduce the memory usage)
the path to the output directory
the project name
the path to the Metagenome Assembled Contig fasta file (FNA)
the path to the Sam/Bam file from read mapping back to contigs (SAM.gz or BAM)
the path to contigs functional annotation result (GFF)
the path to the text file which contains mapping of headers between SAM or BAM and GFF (ID in SAM/FNA<tab>ID in GFF). A two column tab-delimited file. When the annotation and assembly are performed using different identifiers for contigs. The map file is to link the gff file content and mapping result bam file content to the assembled contigs ID.
the path to the database directory which includes checkM_DB and GTDBTK_DB subdirectories.
(optional) scratch_dir: use –scratch_dir for gtdbtk disk swap to reduce memory usage but longer runtime
An example JSON file is shown below:
{
"nmdc_mags.cpu":32,
"nmdc_mags.pplacer_cpu":1,
"nmdc_mags.outdir":"/path/to/output",
"nmdc_mags.proj_name":" Ga0482263",
"nmdc_mags.contig_file":"/path/to/Ga0482263_contigs.fna ",
"nmdc_mags.sam_file":"/path/to/pairedMapped_sorted.bam ",
"nmdc_mags.gff_file":"/path/to/Ga0482263_functional_annotation.gff",
"nmdc_mags.map_file":"/path/to/Ga0482263_contig_names_mapping.tsv",
"nmdc_mags.gtdbtk_database":"/path/to/GTDBTK_DB"
"nmdc_mags.scratch_dir":"/path/to/scratch_dir"
}
Output
The workflow creates several output directories with many files. The main output files, the binned contig files from HQ and MQ bins, are in the hqmq-metabat-bins directory; the corresponding lineage results for the HQ and MQ bins are in the gtdbtk_output directory.
A partial JSON output file is shown below:
|-- MAGs_stats.json
|-- 3300037552.bam.sorted
|-- 3300037552.depth
|-- 3300037552.depth.mapped
|-- bins.lowDepth.fa
|-- bins.tooShort.fa
|-- bins.unbinned.fa
|-- checkm-out
| |-- bins/
| |-- checkm.log
| |-- lineage.ms
| `-- storage
|-- checkm_qa.out
|-- gtdbtk_output
| |-- align/
| |-- classify/
| |-- identify/
| |-- gtdbtk.ar122.classify.tree -> classify/gtdbtk.ar122.classify.tree
| |-- gtdbtk.ar122.markers_summary.tsv -> identify/gtdbtk.ar122.markers_summary.tsv
| |-- gtdbtk.ar122.summary.tsv -> classify/gtdbtk.ar122.summary.tsv
| |-- gtdbtk.bac120.classify.tree -> classify/gtdbtk.bac120.classify.tree
| |-- gtdbtk.bac120.markers_summary.tsv -> identify/gtdbtk.bac120.markers_summary.tsv
| |-- gtdbtk.bac120.summary.tsv -> classify/gtdbtk.bac120.summary.tsv
| `-- ..etc
|-- hqmq-metabat-bins
| |-- bins.11.fa
| |-- bins.13.fa
| `-- ... etc
|-- mbin-2020-05-24.sqlite
|-- mbin-nmdc.20200524.log
|-- metabat-bins
| |-- bins.1.fa
| |-- bins.10.fa
| `-- ... etc
Below is an example of all the output directory files with descriptions to the right.
FileName/DirectoryName |
Description |
|---|---|
1781_86104.bam.sorted |
sorted input bam file |
1781_86104.depth |
the contig depth coverage |
1781_86104.depth.mapped |
the name mapped contig depth coverage |
MAGs_stats.json |
MAGs statistics in json format |
bins.lowDepth.fa |
lowDepth (mean cov <1 ) filtered contigs fasta file by metaBat2 |
bins.tooShort.fa |
tooShort (< 3kb) filtered contigs fasta file by metaBat2 |
bins.unbinned.fa |
unbinned fasta file |
metabat-bins/ |
initial metabat2 binning result fasta output directory |
checkm-out/bins/ |
hmm and marker genes analysis result directory for each bin |
checkm-out/checkm.log |
checkm run log file |
checkm-out/lineage.ms |
lists the markers used to assign taxonomy and the taxonomic level to which the bin |
checkm-out/storage/ |
intermediate file directory |
checkm_qa.out |
checkm statistics report |
hqmq-metabat-bins/ |
HQ and MQ bins contigs fasta files directory |
gtdbtk_output/identify/ |
gtdbtk marker genes identify result directory |
gtdbtk_output/align/ |
gtdbtk genomes alignment result directory |
gtdbtk_output/classify/ |
gtdbtk genomes classification result directory |
gtdbtk_output/gtdbtk.ar122.classify.tree |
archaeal reference tree in Newick format containing analyzed genomes (bins) |
gtdbtk_output/gtdbtk.ar122.markers_summary.tsv |
summary tsv file for gtdbtk marker genes identify from the archaeal 122 marker set |
gtdbtk_output/gtdbtk.ar122.summary.tsv |
summary tsv file for gtdbtk archaeal genomes (bins) classification |
gtdbtk_output/gtdbtk.bac120.classify.tree |
bacterial reference tree in Newick format containing analyzed genomes (bins) |
gtdbtk_output/gtdbtk.bac120.markers_summary.tsv |
summary tsv file for gtdbtk marker genes identify from the bacterial 120 marker set |
gtdbtk_output/gtdbtk.bac120.summary.tsv |
summary tsv file for gtdbtk bacterial genomes (bins) classification |
gtdbtk_output/gtdbtk.bac120.filtered.tsv |
a list of genomes with an insufficient number of amino acids in MSA |
gtdbtk_output/gtdbtk.bac120.msa.fasta |
the MSA of the user genomes (bins) and the GTDB genomes |
gtdbtk_output/gtdbtk.bac120.user_msa.fasta |
the MSA of the user genomes (bins) only |
gtdbtk_output/gtdbtk.translation_table_summary.tsv |
the translation table determined for each sgenome (bins) |
gtdbtk_output/gtdbtk.warnings.log |
gtdbtk warning message log |
mbin-2021-01-31.sqlite |
sqlite db file stores MAGs metadata and statistics |
mbin-nmdc.20210131.log |
the mbin-nmdc pipeline run log file |
rc |
cromwell script sbumit return code |
script |
Task run commands |
script.background |
Bash script to run script.submit |
script.submit |
cromwell submit commands |
stderr |
standard error where task writes error message to |
stderr.background |
standard error where bash script writes error message to |
stdout |
standard output where task writes error message to |
stdout.background |
standard output where bash script writes error message to |
complete.mbin |
the dummy file to indicate the finish of the pipeline |
Version History
1.0.4 (release date 01/12/2022; previous versions: b1.0.3)
Point of contact
Original author: Neha Varghese <njvarghese@lbl.gov>
Package maintainer: Chienchi Lo <chienchi@lanl.gov>
Metatranscriptome Workflow (v0.0.2)
Summary
MetaT is a workflow designed to analyze metatranscriptomes, building on top of already existing NMDC workflows for processing input. The metatranscriptoimics workflow takes in raw data and starts by quality filtering the reads using the RQC worfklow. With filtered reads, the workflow filters out rRNA reads (and separates the interleaved file into separate files for the pairs) using bbduk (BBTools). After the filtering steps, reads are assembled into transcripts and using MEGAHIT and annotated using the Metagenome Anotation Workflow; producing GFF funtional annotation files. Features are counted with Subread’s featureCounts which assigns mapped reads to genomic features and generating RPKMs for each feature in a GFF file for sense and antisense reads.
Workflow Diagram
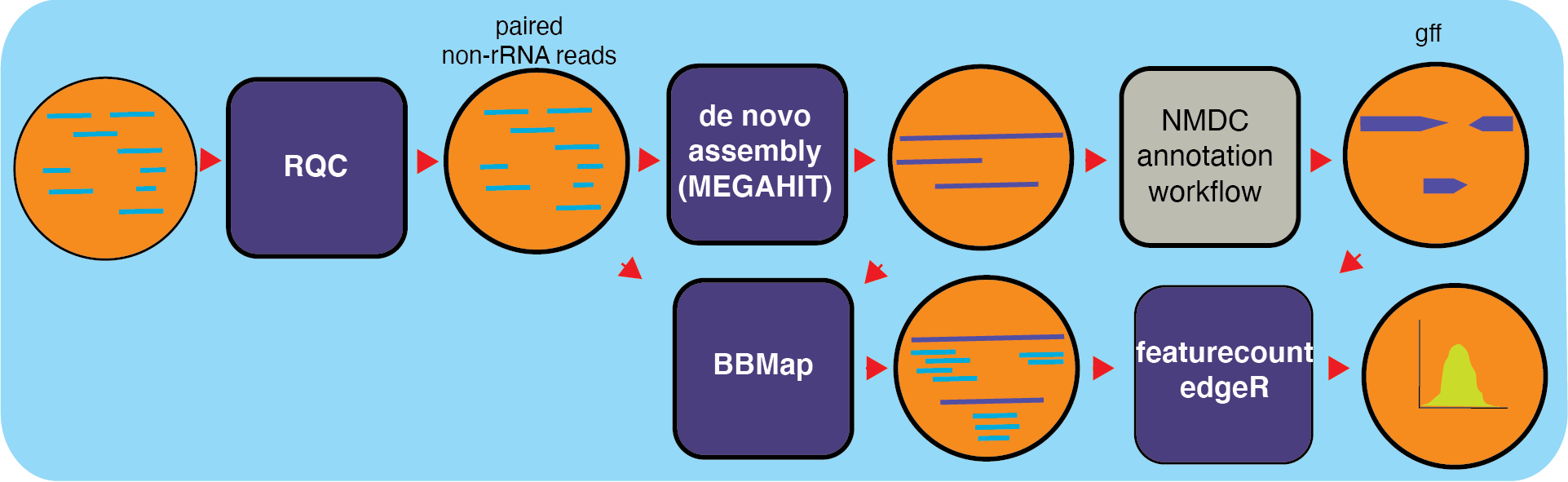
Workflow Availability
The workflow uses the listed docker images to run all third-party tools. The workflow is available in GitHub: https://github.com/microbiomedata/metaT; and the corresponding Docker images that have all the required dependencies are available in following DockerHub (https://hub.docker.com/r/microbiomedata/bbtools, https://hub.docker.com/r/microbiomedata/meta_t, and https://hub.docker.com/r/intelliseqngs/hisat2)
Requirements for Execution (recommendations are in bold):
WDL-capable Workflow Execution Tool (Cromwell)
Container Runtime that can load Docker images (Docker v2.1.0.3 or higher)
Workflow Dependencies
Third-party software (These are included in the Docker images.)
BBTools v38.94. (License: BSD-3-Clause-LBNL.)
BBMap v38.94. (License: BSD-3-Clause-LBNL.)
Python v3.7.6. (License: Python Software Foundation License)
featureCounts v2.0.2. (License: GNU-GPL)
R v3.6.0. (License: GPL-2/GPL-3)
edgeR v3.28.1. (R package) (License: GPL (>=2))
pandas v1.0.5. (python package) (License: BSD-3-Clause)
gffutils v0.10.1. (python package) (License: MIT)
Requisite database
The RQCFilterData Database must be downloaded and installed. This is a 106 GB tar file which includes reference datasets of artifacts, adapters, contaminants, the phiX genome, rRNA kmers, and some host genomes. The following commands will download the database:
wget http://portal.nersc.gov/dna/microbial/assembly/bushnell/RQCFilterData.tar
tar -xvf RQCFilterData.tar
rm RQCFilterData.tar
Sample dataset(s)
The following files are provided with the GitHub download in the test_data folder:
Raw reads: test_data/test_interleave.fastq.gz (output from ReadsQC workflow)
Annotation file: test_functional_annotation.gff (output from mg_annotation workflow)
Input: A JSON file containing the following
a name for the analysis
the number of cpus requested
the path to the clean input interleaved fastq file (recommended: the output from the Reads QC workflow)
the path to the rRNA_kmer database provided as part of RQCFilterData
the path to the assembled transcripts (output of part 1)
the paths to the reads with rRNA removed (paired-end files) (output of part 1)
the path to the annotation file (from the Metagenome Annotation workflow)
An example JSON file is shown below:
{
"metat_omics.project_name": "test",
"metat_omics.no_of_cpus": 1,
"metat_omics.rqc_clean_reads": "test_data/test_interleave.fastq",
"metat_omics.ribo_kmer_file": "/path/to/riboKmers20fused.fa.gz",
"metat_omics.metat_contig_fn": "/path/to/megahit_assem.contigs.fa",
"metat_omics.non_ribo_reads": [
"/path/to/filtered_R1.fastq",
"/path/to/filtered_R2.fastq"
],
"metat_omics.ann_gff_fn": "test_data/test_functional_annotation.gff"
}
Output
Output is split up between steps of the workflow. The first half of the workflow will output rRNA-filtered reads and the assembled transcripts. After annotations and featureCount steps include a JSON file that contain RPKMs for both sense and antisense, reads, and information from annotation for each feature. An example of JSON outpus:
{
"featuretype": "transcript",
"seqid": "k123_15",
"id": "STRG.2.1",
"source": "StringTie",
"start": 1,
"end": 491,
"length": 491,
"strand": ".",
"frame": ".",
"extra": [],
"cov": "5.928717",
"FPKM": "76638.023438",
"TPM": "146003.046875"
}
Below is an example of the output directory files with descriptions to the right.
Directory/File Name |
Description |
|---|---|
metat_output/sense_out.json |
RPKM for each feature on + strand |
metat_output/antisense_out.json |
RPKM for each feature on - strand |
assembly/megahit_assem.contigs.fa |
assembled transcripts |
mapback/mapped_sorted.bam |
alignment of reads and transcripts |
qa/_interleaved.fastq |
non-ribosomal reads |
qa/filterStats.txt |
summary statistics in JSON format |
qa/filterStats2.txt |
more detailed summary statistics |
annotation/annotations.json |
annotation information |
annotation/features.json |
feature information |
annotation/_cath_funfam.gff |
features from cath database |
annotation/_cog.gff |
features from cog databse |
annotation/_ko_ec.gff |
features from ko database |
annotation/_pfam.gff |
features from pfam database |
annotation/_smart.gff |
features from smart database |
annotation/_structural_annotation.gff |
structural features |
annotation/_supfam.gff |
features from supfam databse |
annotation/_tigrfam.gff |
features from trigfam database |
annotation/_functional_annotation.gff |
functional features |
annotation/_ec.tsv |
ec terms tsv |
annotation/_ko.tsv |
ko terms tsv |
annotation/proteins.faa |
fasta containing protiens |
Version History
0.0.2 (release date 01/14/2021; previous versions: 0.0.1)
0.0.3 (release date 07/28/2021; previous versions: 0.0.2)
Points of contact
Author: Migun Shakya <migun@lanl.gov>
Metaproteomic Workflow (v1.0.0)
Summary
The metaproteomics workflow/pipeline is an end-to-end data processing workflow for protein identification and characterization using MS/MS data. Briefly, mass spectrometry instrument generated data files(.RAW) are converted to mzML, an open data format, using MSConvert. Peptide identification is achieved using MSGF+ and the associated metagenomic information in the FASTA (protein sequences) file format. Intensity information for identified species is extracted using MASIC and combined with protein information.
Workflow Diagram
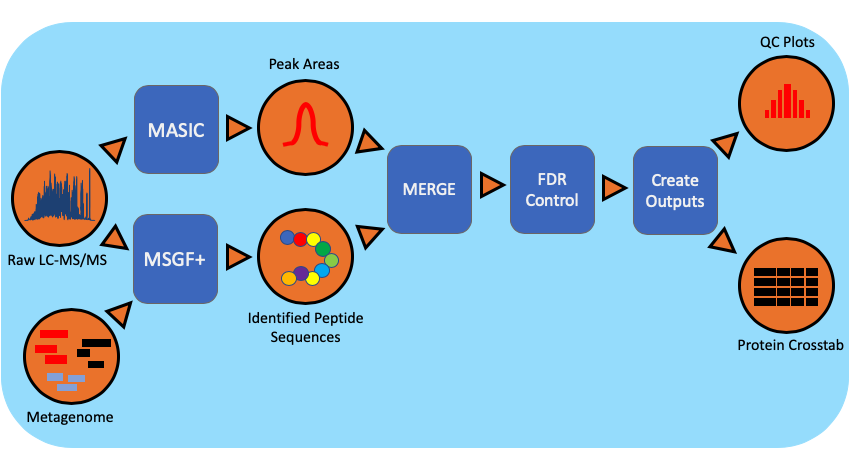
Workflow Dependencies
Third party software
|----------------------------|------------------------------------------|
| MSGFPlus | v20190628 |
| Mzid-To-Tsv-Converter | v1.3.3 |
| PeptideHitResultsProcessor | v1.5.7130 |
| pwiz-bin-windows | x86_64-vc141-release-3_0_20149_b73158966 |
| MASIC | v3.0.7235 |
| sqlite-netFx-full-source | 1.0.111.0 |
| Conda | (3-clause BSD) |
| | |
Workflow Availability
The workflow is available in GitHub: https://github.com/microbiomedata/metaPro
The container is available at Docker Hub (microbiomedata/mepro): https://hub.docker.com/r/microbiomedata/mepro
Inputs
.raw, metagenome, parameter files : MSGFplus & MASIC, contaminant_file
Outputs
Processing multiple datasets.
.
├── Data/
├── FDR_table.csv
├── Plots/
├── dataset_job_map.csv
├── peak_area_crosstab_by_dataset_id.csv
├── protein_peptide_map.csv
├── specID_table.csv
└── spectra_count_crosstab_by_dataset_id.csv
Processing single FICUS dataset.
metadatafile, [Example](https://jsonblob.com/400362ef-c70c-11ea-bf3d-05dfba40675b)
<pre><code>| Keys | Values | |--------------------|————————————————————————–| | id | str: “md5 hash of $github_url+$started_at_time+$ended_at_time” | | name | str: “Metagenome:$proposal_extid_$sample_extid:$sequencing_project_extid | | was_informed_by | str: “GOLD_Project_ID” | | started_at_time | str: “metaPro start-time” | | ended_at_time | str: “metaPro end-time” | | type | str: tag: “nmdc:metaPro” | | execution_resource | str: infrastructure name to run metaPro | | git_url | str: “url to a release” | | dataset_id | str: “dataset’s unique-id at EMSL” | | dataset_name | str: “dataset’s name at EMSL” | | has_inputs | json_obj | | has_outputs | json_obj | | stats | json_obj | </code></pre>
has_inputs : | MSMS_out | str: file_name |file_size |checksum | | metagenome_file | str: file_name |file_size |checksum |
int: entry_count(#of gene sequences) | int: duplicate_count(#of duplicate gene sequences) |
parameter_files | str: for_masic/for_msgfplus : file_name |file_size |checksum parameter file used for peptide identification searchContaminant_file | str: file_name |file_size |checksum (FASTA containing common contaminants in proteomics)has_outputs: | collapsed_fasta_file | str: file_name |file_size |checksum | | resultant_file | str: file_name |file_size |checksum | | data_out_table | str: file_name |file_size |checksum |
stats: | from_collapsed_fasta | int: entry_count(#of unique gene sequences) | | from_resultant_file | int: total_protein_count | | from_data_out_table | int: PSM(# of MS/MS spectra matched to a peptide sequence at 5% false discovery rate (FDR)
float: PSM_identification_rate(# of peptide matching MS/MS spectra divided by total spectra searched (5% FDR) int: unique_peptide_seq_count(# of unique peptide sequences observed in pipeline analysis 5% FDR) int: first_hit_protein_count(# of proteins observed assuming single peptide-to-protein relationships) int: mean_peptide_count(Unique peptide sequences matching to each identified protein.)
data_out_table
| DatasetName | PeptideSequence | FirstHitProtein | SpectralCount | sum(MasicAbundance) | GeneCount | FullGeneList | FirstHitDescription | DescriptionList | min(Qvalue) |
collapsed_fasta_file
resultant_file
Requirements for Execution
Docker or other Container Runtime
Version History
1.0.0
Point of contact
Package maintainer: Anubhav <anubhav@pnnl.gov>
Metabolomics Workflow
Summary
The gas chromatography-mass spectrometry (GC-MS) based metabolomics workflow (metaMS) has been developed by leveraging PNNL’s CoreMS software framework. The current software design allows for the orchestration of the metabolite characterization pipeline, i.e., signal noise reduction, m/z based Chromatogram Peak Deconvolution, abundance threshold calculation, peak picking, spectral similarity calculation and molecular search, similarity score calculation, and confidence filtering, all in a single step.
Workflow Diagram
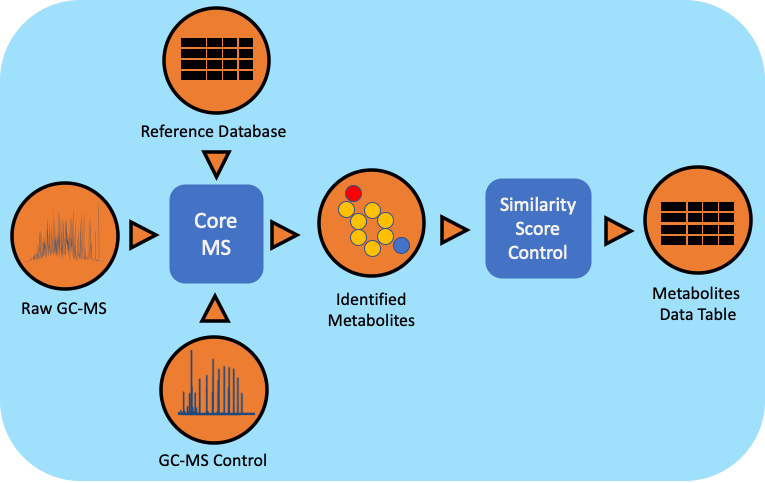
Workflow Dependencies
Third party software
CoreMS (2-clause BSD)
Click (BSD 3-Clause “New” or “Revised” License)
Database
PNNL Metabolomics GC-MS Spectral Database
Workflow Availability
The workflow is available in GitHub: https://github.com/microbiomedata/metaMS
The container is available at Docker Hub (microbiomedata/metaMS): https://hub.docker.com/r/microbiomedata/metams
The python package is available on PyPi: https://pypi.org/project/metaMS/
The databases are available by request. Please contact NMDC (support@microbiomedata.org) for access.
Test datasets
https://github.com/microbiomedata/metaMS/blob/master/data/GCMS_FAMES_01_GCMS-01_20191023.cdf
Execution Details
Please refer to:
https://github.com/microbiomedata/metaMS#metams-installation
Inputs
- Supported format for low resolution GC-MS data:
ANDI NetCDF for GC-MS (.cdf)
- Fatty Acid Methyl Esters Calibration File:
ANDI NetCDF for GC-MS (.cdf) - C8 to C30
- Parameters:
CoreMS Parameter File (.json)
MetaMS Parameter File (.json)
Outputs
- Metabolites data-table
CSV, TAB-SEPARATED TXT
HDF: CoreMS HDF5 format
XLSX : Microsoft Excel
- Workflow Metadata:
JSON
Requirements for Execution
Docker Container Runtime
or
Python Environment >= 3.6
Python Dependencies are listed on requirements.txt
Version History
2.1.3
Point of contact
Package maintainer: Yuri E. Corilo <corilo@pnnl.gov>
Natural Organic Matter Workflow
Summary
Direct Infusion Fourier Transform mass spectrometry (DI FT-MS) data undergoes signal processing and molecular formula assignment leveraging EMSL’s CoreMS framework. Raw time domain data is transformed into the m/z domain using Fourier Transform and Ledford equation. Data is denoised followed by peak picking, recalibration using an external reference list of known compounds, and searched against a dynamically generated molecular formula library with a defined molecular search space. The confidence scores for all the molecular formula candidates are calculated based on the mass accuracy and fine isotopic structure, and the best candidate assigned as the highest score.
Workflow Diagram
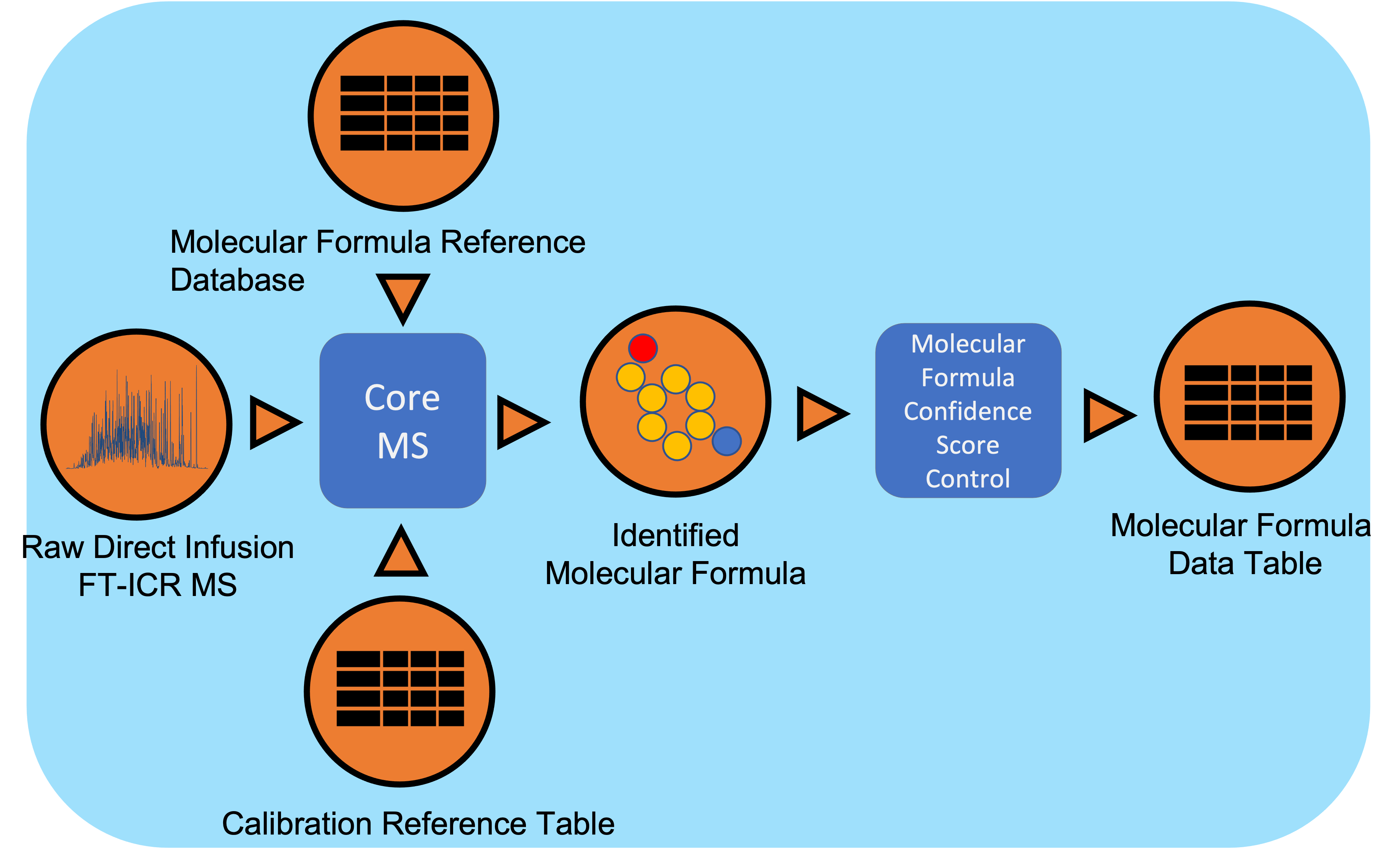
Workflow Dependencies
Third party software
CoreMS (2-clause BSD)
Click (BSD 3-Clause “New” or “Revised” License)
Database
CoreMS dynamic molecular database search and generator
Workflow Availability
The workflow is available in GitHub: https://github.com/microbiomedata/enviroMS
The container is available at Docker Hub (microbiomedata/metaMS): https://hub.docker.com/r/microbiomedata/enviroms
The python package is available on PyPi: https://pypi.org/project/enviroMS/
The databases are available by request. Please contact NMDC (support@microbiomedata.org) for access.
Test datasets
Execution Details
Please refer to:
https://github.com/microbiomedata/enviroMS#enviroms-installation
Inputs
- Supported format for Direct Infusion FT-MS data:
Thermo raw file (.raw)
Bruker raw file (.d)
Generic mass list in profile and/or centroid mode (inclusive of all delimiters types and Excel formats)
- Calibration File:
Molecular Formula Reference (.ref)
- Parameters:
CoreMS Parameter File (.json)
EnviroMS Parameter File (.json)
Outputs
- Molecular Formula Data-Table, containing m/z measuments, Peak height, Peak Area, Molecular Formula Identification, Ion Type, Confidence Score, etc.
CSV, TAB-SEPARATED TXT
HDF: CoreMS HDF5 format
XLSX : Microsoft Excel
- Workflow Metadata:
JSON
Requirements for Execution
Docker Container Runtime or
Python Environment >= 3.8 and
Python Dependencies are listed on requirements.txt
Version History
4.1.5
Point of contact
Package maintainer: Yuri E. Corilo <corilo@pnnl.gov>